DNase I Footprinting Assay Service
Background
Introduction
Understanding how DNA and proteins interact is key to figuring out gene regulation, transcription control, and many biological processes. These interactions are vital because transcription factors attach to specific DNA spots to adjust how genes are expressed. Pinpointing exactly where these proteins bind is crucial for digging into the molecular workings of cellular functions and diseases. Traditional techniques like ChIP-seq and EMSA have their downsides, especially when it comes to resolution and throughput. The DNase I footprinting assay is a fantastic alternative, offering high-resolution details about protein-DNA interactions, making it a must-have for researchers in molecular biology and genetics. Profacgen's DNase I footprinting assay service is crafted to help researchers tackle these hurdles, providing a thorough and sensitive way to spot and study DNA-protein interactions.
What is DNase I Footprinting Assay?
This approach involves using DNase I to cut DNA fragments that are either radioactively or fluorescently labeled. Gel electrophoresis is then applied to show the cutting pattern. You compare the pattern of DNA without any binding proteins, known as free DNA, to the pattern when a DNA-binding protein is there. If the protein attaches to the DNA, it shields that spot from being cut by the enzyme, leaving a clear "footprint" on the gel. By adjusting the protein concentration, you can gauge its binding strength based on the smallest amount needed to create a footprint.
Traditionally, DNase I footprinting involves radioactive labeling of DNA and running a sequencing gel for detection. However, due to safety issues and the inconvenience of radioactive labels, Profacgen opts for fluorescence labeling. Using this along with a sequencer, like those from Applied Biosystem, makes our DNase I footprinting assay safer, quicker, and more sensitive.
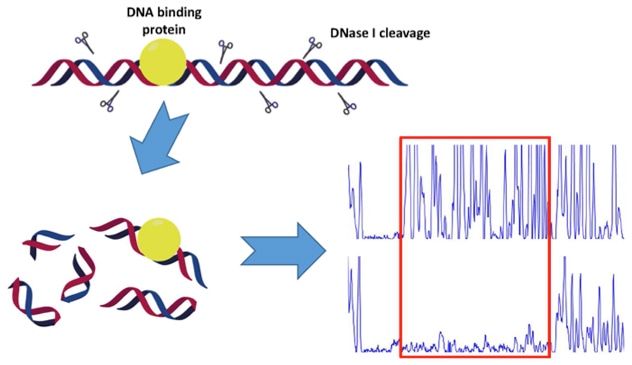 Fig1. Schematic Diagram of the DNase I Footprinting Assay.
Fig1. Schematic Diagram of the DNase I Footprinting Assay.
Applications of DNase I Footprinting Assay
The DNase I footprinting assay is used a lot in molecular biology and genetics. It's especially handy for:
- Finding Transcription Factor Binding Sites: Pinpointing exactly where transcription factors attach to DNA sequences.
- Mapping Regulatory Elements: Figuring out regulatory spots in gene promoters and enhancers to grasp how genes are controlled.
- Validating Drug Targets: Checking potential drug targets by identifying proteins that bind with certain DNA sequences.
- Functional Annotation: Offering detailed annotations of DNA sequences by spotting areas that avoid DNase I cutting.
Service Procedure
As DNase I footprinting assay is used to determine the precise DNA sequence bound by the protein of interest, it is recommended to be performed after interaction confirmation by Electrophoretic Mobility Shift Assay (EMSA). Therefore, in addition to purified protein and the DNA fragment, EMSA data and relating experimental parameters will all help design the DNase I footprinting assay.
Deliverables:
- High-resolution DNA-protein interaction data.
- Detailed analysis reports, including binding site identification and affinity estimation.
- Visualizations of cleavage patterns and binding sites.

What We Offer?
Sample Prep and DNA Labeling
We start by carefully preparing your samples to make sure we have top-notch DNA fragments and proteins. We can handle the DNA fragments and protein prep if you need. We then label one end of the DNA with either a radioactive or fluorescent tag to clearly see where proteins bind.
Protein-DNA Binding and DNase I Digestion
After labeling, we mix the DNA with the protein in a special buffer, letting them bind under controlled conditions. We then use DNase I to partially cut the DNA. This step reveals which parts of the DNA are protected by the protein, creating the "footprint."
DNA Purification and Gel Electrophoresis
Post-digestion, we clean up the DNA to get rid of any leftover proteins or contaminants. The purified DNA is run on a gel to separate it by size, showing the protected areas as clear bands. This high-resolution image shows exactly where proteins bind.
Data Analysis and Result Interpretation
Finally, we analyze the labeled DNA fragments using imaging techniques. Our team examines the cleavage patterns to find protected regions, or "footprints," which point to specific binding sites. We also determine the protein's binding strength by adjusting its concentration and noting the footprint intensity. You'll get detailed reports with visuals and insights into DNA-protein interactions.
Why Choose Profacgen?
- Great Sensitivity and Consistency: Our DNase I footprinting assay provides highly sensitive and consistent results, making sure you accurately find the binding sites.
- Skilled Technicians: Our skilled technicians make sure you get top-quality and dependable data.
- Tailored Experimental Design: We offer customized experimental designs to meet your specific research needs.
- Best Price in the Market: Competitive pricing without compromising on quality or service.
Case Study
Project: DNase I Footprinting Assay for X Protein
Goals
The aim of the project is to identify the DNA binding sites of XXX protein using DNase I footprinting assay. Briefly, one oligo was designed and synthesized in vitro, labeled with FAM. After incubation with purified X protein, the complexes were digested with DNase I, and the purified DNA was subjected to sequencing.
Methods and Materials
- Samples: 500 μg X proteins were provided by the customer.
- Equipment:
- PCR Clean-Up System
- NanoDrop 2000C
- ABI sequencer
Results
When 250 ng probes were incubated with 10 µg of protein X, an obvious protein protection region was observed, covering about 77 bases. The results indicate an interaction between X protein and the synthesized oligo. The binding sites of X protein with its target DNA were identified as specific sequences, providing insights into the mechanism of gene regulation.
FAQs
Resources
References:
- Cardew AS, Fox KR. DNase I footprinting. Methods Mol Biol. 2010;613:153-172.
- Sivapragasam S, Pande A, Grove A. A recommended workflow for DNase I footprinting using a capillary electrophoresis genetic analyzer. Anal Biochem. 2015;481:1-3.