Membrane-Based Yeast Two-Hybrid Screening
Background
Introduction
Membrane proteins are central to cellular functions such as signal transduction, transport, and intercellular communication, yet studying their interactions remains challenging due to their hydrophobic nature and dependence on lipid environments. Traditional methods like co-IP or FRET often struggle with solubility issues and fail to mimic native membrane conditions. The Membrane-based Yeast Two-Hybrid (MbY2H) system overcomes these limitations by enabling interaction analysis within an authentic membrane context, preserving protein conformations and modifications critical for functionality.
As membrane proteins represent over 60% of therapeutic targets, MbY2H is pivotal in drug discovery, particularly for diseases linked to receptors, channels, and transporters.
Advance Your Research With Our Service
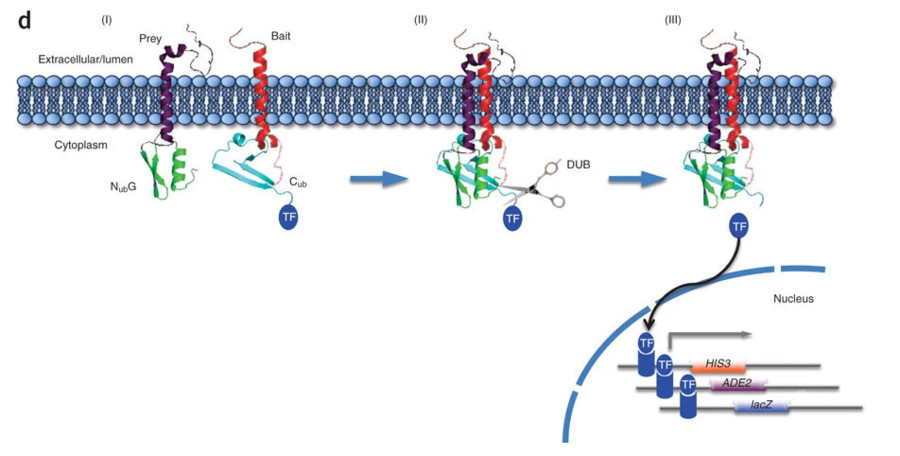 Fig1. Important elements of the MYTH system. (Snider, et al., 2010)
Fig1. Important elements of the MYTH system. (Snider, et al., 2010)
How It Works
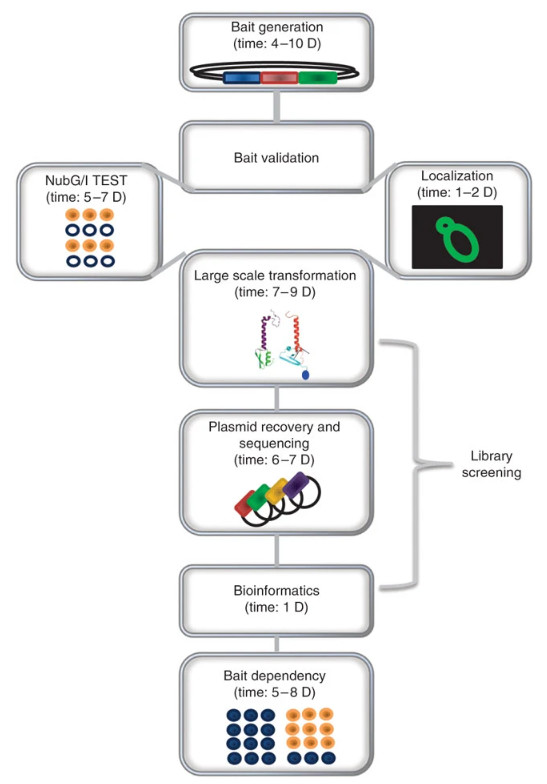 Fig2. The MYTH pipeline. (Snider, et al., 2010)
Fig2. The MYTH pipeline. (Snider, et al., 2010)
1. Principle of the Split-Ubiquitin System
The MbY2H system utilizes a split-ubiquitin complementation assay. Cub (C-terminal ubiquitin) and mutated Nub (N-terminal ubiquitin) fragments reconstitute functional ubiquitin only when bait-prey interactions bring them into proximity, enabling precise detection of membrane protein interactions.
2. Bait and Prey Protein Fusion
The bait (membrane protein) is fused to Cub and an artificial transcription factor (TF), while the prey is linked to a low-affinity Nub. This design minimizes spontaneous Cub-Nub binding, ensuring interaction-dependent ubiquitin reassembly and reducing false positives.
3. Transcription Factor Release and Reporter Activation
Ubiquitin reconstitution recruits proteases to cleave the TF from the bait. The released TF enters the nucleus, activating reporter genes (e.g., HIS3) for selective growth or enzymatic readouts, enabling high-throughput interaction screening.
4. Visual Workflow Overview
Key steps include cloning bait/prey into Cub-TF/Nub vectors, co-transforming yeast, selective culture to eliminate non-interactors, and quantifying interactions via reporter activation-ideal for hydrophobic or low-abundance membrane targets.
5. Technical Advantages
The system operates in live yeast, preserving native membrane contexts and post-translational modifications. It supports studies on GPCRs, transporters, and disease-related targets, bridging functional proteomics and drug discovery.
Applications of Our Service
| Types | Key Point | Description |
| Drug Target Identification | Identifying novel drug targets through membrane protein interactions. | Identify potential drug targets by screening interactions of membrane proteins. |
| Pathway Elucidation | Mapping signaling pathways involving membrane proteins. | Map signaling pathways by identifying interaction networks of membrane proteins. |
| Disease Research | Investigating the role of membrane proteins in disease mechanisms. | Study disease mechanisms by exploring interactions of disease-related membrane proteins. |
| Agricultural Research | Enhancing crop traits through membrane protein interactions. | Improve crop traits by identifying interactions of membrane proteins. |
| Functional Genomics | Exploring the interactome of membrane proteins in various organisms. | Use MbY2H to identify large-scale interaction networks of membrane proteins. |
| High-Throughput Screening | Efficient screening of membrane protein interactions. | Screen large cDNA libraries to identify novel membrane protein interactions. |
Service Procedure
Sample Submission Requirements
We're excited to help you explore protein interactions with our Membrane-Based Yeast Two-Hybrid Screening service! To get started smoothly, we kindly ask for the following information and samples:
- Target Protein Information
Please provide the gene sequence of your target protein, including the cDNA sequence. If you have any background information or specific requirements, feel free to share them with us. This will help us tailor our service to your needs. - Bait Vector (Optional)
If you already have a bait vector constructed, please send it along. If not, don't worry-we can build it for you! Just provide the necessary gene fragment details for primer design. - cDNA Library Selection
We offer a range of standardized cDNA libraries for you to choose from. If you have a custom library, please let us know-we're happy to work with that too! - Sample Quality
For DNA samples, please ensure they are high-purity with a concentration of ≥50 ng/µL. If you're submitting protein samples, make sure they are active and pure. If you have any doubts, feel free to reach out-we're here to help! - Additional Details
Any extra information about your target protein, such as known interactions or specific screening conditions, will be very helpful for us to optimize the process.
Delivery
- Detailed Screening Report
- Positive Clone Sequences & BLAST Results
- High-Throughput Sequencing Analysis
- Optional Validation Data (Co-IP, Pull-down)
- Standard Timeline: 8-10 weeks
- Expedited Timeline: 6-8 weeks
Service Scope
| Bait Vector Construction | Library Screening | Self-Activation Test | Positive Clone Identification | Back Transformation and Validation | Customized Solutions |
| Primer design, gene amplification, and cloning validation services for bait vector construction. | High-throughput screening using standardized or custom cDNA libraries. | Self-activation testing of bait vectors to ensure functionality. | Identification of positive clones via PCR, sequencing, and BLAST alignment, with interaction partner information provided. | Back-transformation and validation experiments to ensure result reliability. | Customized services including optimization for special protein screening and full technical support. |
Why Choose Profacgen?
Specialized Focus on Membrane Protein Studies
Our team excels in membrane protein interaction research, utilizing the MbY2H system to tackle challenges like protein solubility and native conformation preservation. This expertise ensures reliable results for targets such as GPCRs and ion channels.
Tailored End-to-End Solutions
We adapt our workflows to your needs—whether analyzing single interactions or large-scale networks. Services span bait design, library screening, and data analysis, supporting diverse species and membrane protein types.
Quality-Driven Affordability
Competitive pricing pairs with stringent quality checks, including triple-validation (genetic, biochemical, and functional) for every interaction. Transparent quotes with no hidden costs let you plan confidently.
Dedicated Partnership at Every Step
A designated project manager guides you from planning to final reporting, offering insights for experimental optimization and troubleshooting. We prioritize aligning outcomes with your research timelines.
Clear Updates and Faster Results
Track progress in real time through our client portal and receive biweekly summaries. Our optimized pipelines deliver data 25% faster than standard industry timelines, accelerating your discoveries.
Case Study
* NOTE: We prioritize confidentiality in our services to safeguard technology and intellectual property for enhanced future value and protection. The following case study has been shared with the client's consent.
Goal
The goal of this project is to detect proteins that interact with the target protein XXX using the Yeast Two-Hybrid (Y2H) assay and deliver the results to the client. The process includes constructing the bait vector, screening the Arabidopsis membrane yeast cDNA library, multiplex reporter gene detection, DNA sequencing, and BLAST alignment analysis of positive clones to identify interaction partners of the XXX protein.
Results
- Gene Amplification
The XXX gene was successfully amplified using the designed primers. The size of the amplified fragment was approximately 800 bp, as shown in the agarose gel electrophoresis.
 Fig3. Two Agarose Gel Electrophoresis of xxx Gene.
Fig3. Two Agarose Gel Electrophoresis of xxx Gene.
- Ligation and Transformation
Sequence analysis confirmed that the XXX gene was correctly cloned into the pPR3-N vector, with the corresponding amino acids in frame. - Self-activation Test
The self-activation assay was performed, and the results indicated that the bait construct did not activate the reporter genes in the absence of interaction.
 Fig4. Self-activation Testing for xxx.
Fig4. Self-activation Testing for xxx.
- Transformation Efficiency for cDNA Library Screening
The transformation efficiency was evaluated by counting the number of colonies on the SD-TLH plate. A total of 96 monoclonal colonies were observed.
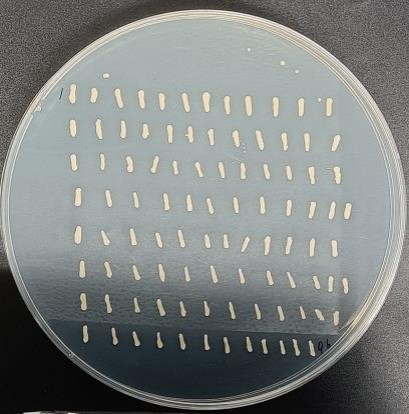 Fig5. Transformation Efficiency for cDNA Library Screening.
Fig5. Transformation Efficiency for cDNA Library Screening.
- Positive Clones Selection
Out of the 96 positive yeast clones, 30 unique sequences were identified through PCR amplification and BLAST analysis. - Back Transformation
The 30 yeast-positive clones were validated by back-transformation and growth on SD-TL, SD-TLH, and SD-TLHA-deficient plates.
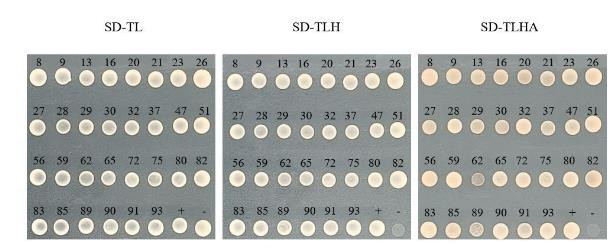 Fig6. Positive Clones Validation Back Transformation Experiment.
Fig6. Positive Clones Validation Back Transformation Experiment.
- High-throughput Sequencing
The positive clones were sequenced using NGS, and the results confirmed the identified interaction partners.
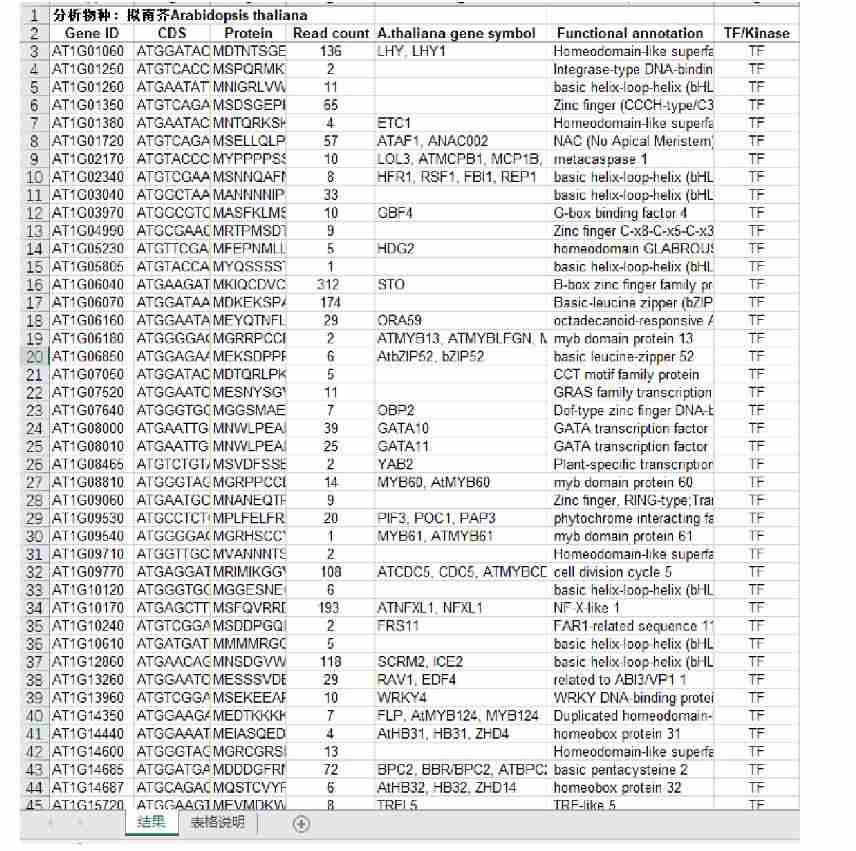 Fig7. High-throughput Sequencing. (partial)
Fig7. High-throughput Sequencing. (partial)
Conclusions and Discussions
Through the experiments, we successfully detected the protein interactions with the target protein XXX using the Y2H assay. We identified 30 unique interaction partners from the screened library and delivered the results to the client. The project demonstrated the effectiveness of the Y2H system in identifying novel protein interactions, particularly for membrane proteins. Future work may include further validation of these interactions using complementary techniques such as co-immunoprecipitation or pull-down assays.
Customer Testimonials
"The MbY2H platform's ability to preserve native protein conformations was critical for our oncology targets. We uncovered GPCR interaction patterns missed by traditional methods-data directly informed two preclinical candidates."
Lead Scientist, Multinational Pharma R&D Division
"Scalability without compromise. Their team processed 450+ membrane transporter interactions in 4 weeks, with 95% validation concordance. This efficiency reshaped our metabolic disease target pipeline."
CTO, Drug Discovery Group
"As a resource-limited team, their flexible payment plans and stepwise validation allowed us to prioritize high-value interactions. "
Dr. Carlos Rivera, Latin American Neurodegeneration Research Hub
"Technical excellence filled our regional expertise gap. Their optimized protocols stabilized lipid raft-dependent receptors our lab failed to analyze for years-now adopted as regional best practices."
Dr. Aisha Al-Mansoori, Middle East Membrane Biology Alliance
"Cross-species compatibility accelerated our platform. Comparing human and primate ion channel interactomes revealed evolutionary-conserved targets, cutting our lead optimization phase by 35%."
Head of R&D, North American Biotech Startup
"Seamless transition from service to in-house adoption. Their MbY2H training modules and 24/7 support enabled our team to replicate cardiac channel studies independently within a month-exceeding expectations."
Dr. Yuki Tanaka, Medicine Research Institute
FAQs
- Genetic reconfirmation in dual selection media.
- Co-purification assays (e.g., Ni-NTA pull-down).
- Functional validation (e.g., ligand-dependent interaction modulation).
A negative control library screen is included by default.
Resources
| Co-Immunoprecipitation (Co-IP) | Surface Plasmon Resonance (SPR) Service | Proximity-dependent Biotin Identification (BioID) Service | Yeast Display Service | Bio-Layer Interferometry (BLI) |
References:
- Snider J.; et al. Split-ubiquitin based membrane yeast two-hybrid (MYTH) system: a powerful tool for identifying protein-protein interactions. J Vis Exp. 2010;(36):1698.
- Snider J.; et al. Detecting interactions with membrane proteins using a membrane two-hybrid assay in yeast. Nat Protoc. 2010;5(7):1281-1293.