ChIRP-Seq Service
Profacgen has accumulated lots of experience in nucleic acid-protein interaction research. Our professional technical team can provide customers with efficient ChIRP-Seq and many related featured services. Our competitive prices and extensive expertise have earned us the trust of our collaborators. Contact us to find out how Profacgen could be of assistance.
<5kb
Spatial resolution
<100 hyperactive cells
Sample size
500+
Species experience
Multiomics integration
Data dimension
Background
Introduction
Non-coding RNAs (ncRNAs), particularly long non-coding RNAs (lncRNAs), are now recognized as master regulators of eukaryotic gene expression. These molecules orchestrate cellular processes-from differentiation to stress responses-by forming dynamic RNA-protein complexes, guiding chromatin modifiers, or acting as scaffolds for transcriptional machinery. Their dysregulation is implicated in pathologies ranging from cancer to neurodegenerative disorders.
ChIRP-seq (Chromatin Isolation by RNA Purification sequencing) has revolutionized our ability to map these interactions in vivo. This technique employs biotinylated probes complementary to target lncRNAs, enabling precise capture of RNA/protein/DNA complexes from crosslinked chromatin. Subsequent streptavidin pulldowns, coupled with RNase digestion and deep sequencing, generate high-resolution maps of RNA-chromatin interactions. Beyond identifying binding sites, ChIRP-seq reveals spatial genome organization and lncRNA-mediated epigenetic states-critical for dissecting disease mechanisms or identifying therapeutic targets.
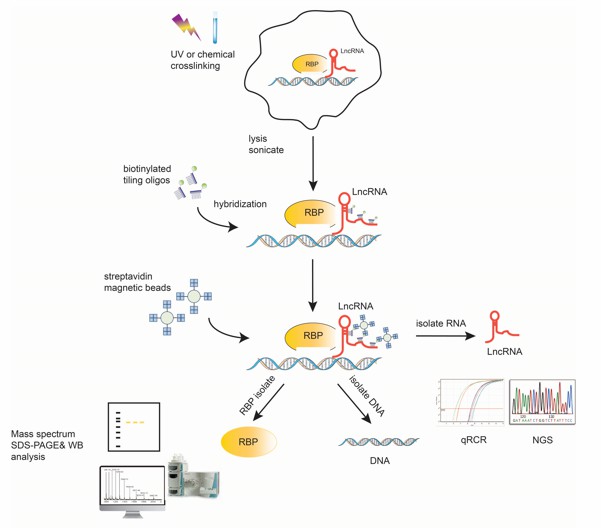 Fig1. The overall scheme of ChIRP-Seq steps in detail.
Fig1. The overall scheme of ChIRP-Seq steps in detail.
Applications of ChIRP-Seq
Disease Research
- Oncology applications: ChIRP-Seq reveals lncRNAs driving cancer growth and metastasis, identifying therapeutic targets.
- Neurodegenerative diseases: This technology uncovers lncRNA roles in neuronal degeneration and survival, aiding treatment research.
- Genetic disorders: ChIRP-Seq helps understand ncRNA contributions to genetic mutations and disease mechanisms.
Developmental Biology
- Embryonic development: ChIRP-Seq tracks ncRNAs guiding cell differentiation and organ formation in embryos.
- Stem cell differentiation: It identifies lncRNAs regulating stem cell fate decisions and lineage commitment.
- Tissue specification: The technology discovers ncRNAs involved in defining specialized tissue identities during development.
Environmental Response
- Stress responses: ChIRP-Seq identifies ncRNAs helping organisms adapt to environmental stressors like heat or toxins.
- Molecular adaptation: It reveals lncRNAs contributing to long-term adaptation of species to ecological changes.
Drug Discovery
- Therapeutic targets: ChIRP-Seq pinpoints ncRNAs suitable for drug development against various diseases.
- Drug mechanisms: The technology clarifies how drugs targeting RNA or chromatin function at the molecular level.
Service Procedure

Our Enhanced ChIRP-Seq Service
| Optimized Protocol | Personalized Solutions | Complementary Services |
|
|
Why Choose Profacgen?
Expertise & Experience: Professional team with rich research experience.
Quality Assurance: Strict quality control in every experiment step.
Customer Support: Full - process technical consultation and assistance.
Fast Turnaround: Efficient processing to accelerate your research.
Custom Solutions: Flexible services tailored to your specific needs.
Case Study
* NOTE: We prioritize confidentiality in our services to safeguard technology and intellectual property for enhanced future value and protection. The following case study has been shared with the client's consent.
Goal
The goal of this project is to screen interaction DNAs of xxx lncRNA in xxx cell lines by ChIRP-Seq. The bound DNAs were analyzed by high-throughput sequencing to identify the genomic binding sites of the lncRNA and explore its potential biological functions through bioinformatics analysis.
Results
- Sequence Alignment, Sequencing Depth and Coverage Statistics
The sequencing data was successfully aligned to the reference genome, with sufficient coverage and depth to ensure reliable detection of RNA-protein interactions.
- Gene Ontology (GO) Enrichment Analysis of Differential Genes
GO enrichment analysis identified significant terms related to the biological processes and functions of the RNA targets, providing insights into their regulatory mechanisms.
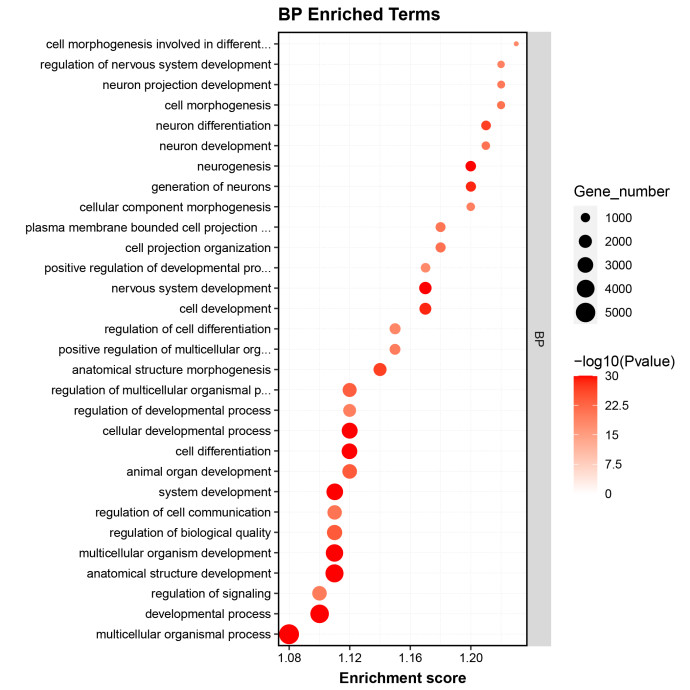
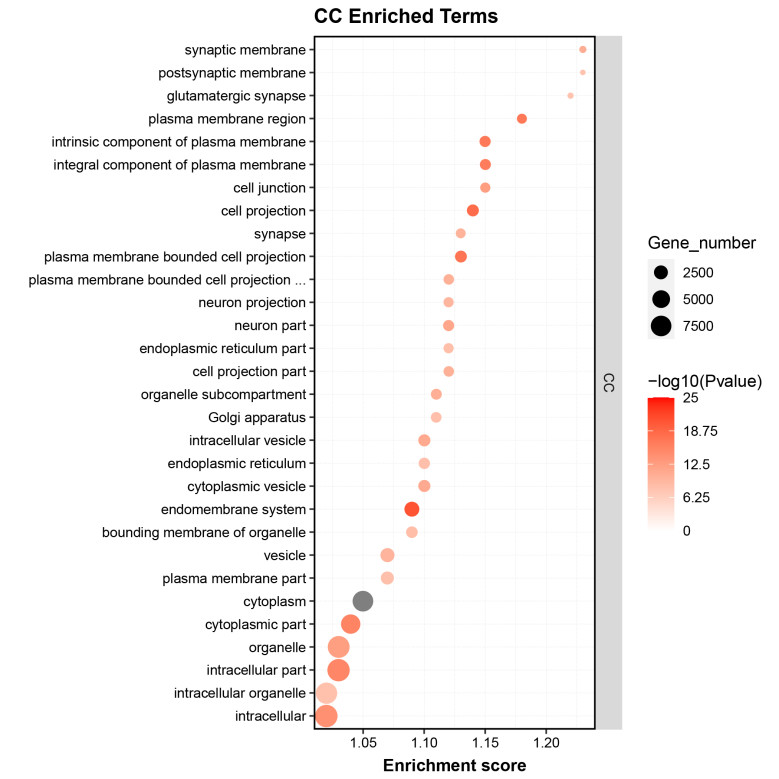
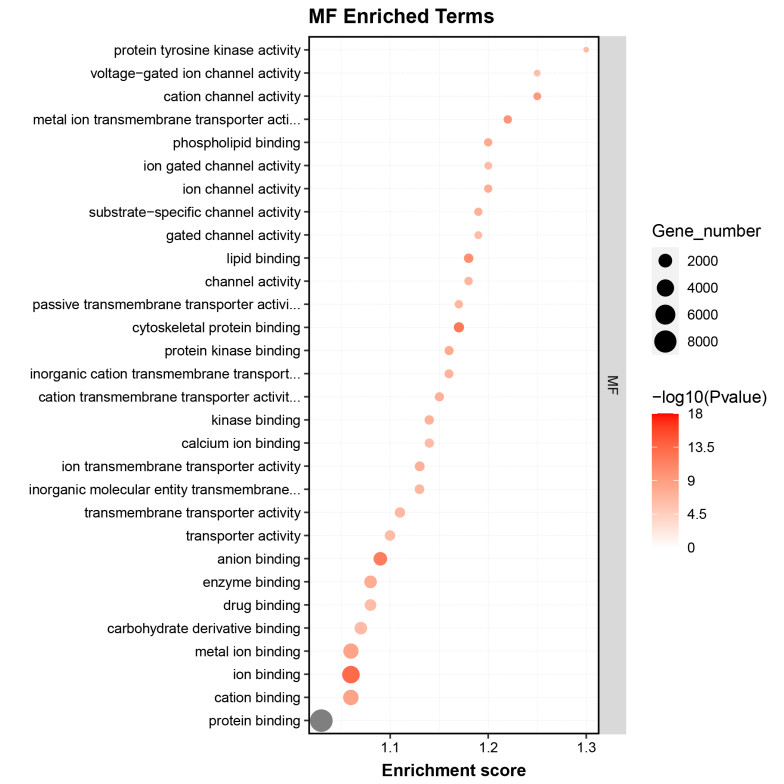
Fig2. GO Enrichment Analysis of Differential Genes.
- KEGG Biological Pathway Analysis of Differential Peak Related Genes
KEGG pathway enrichment analysis revealed key biological pathways associated with the identified RNA targets, highlighting potential functional roles.
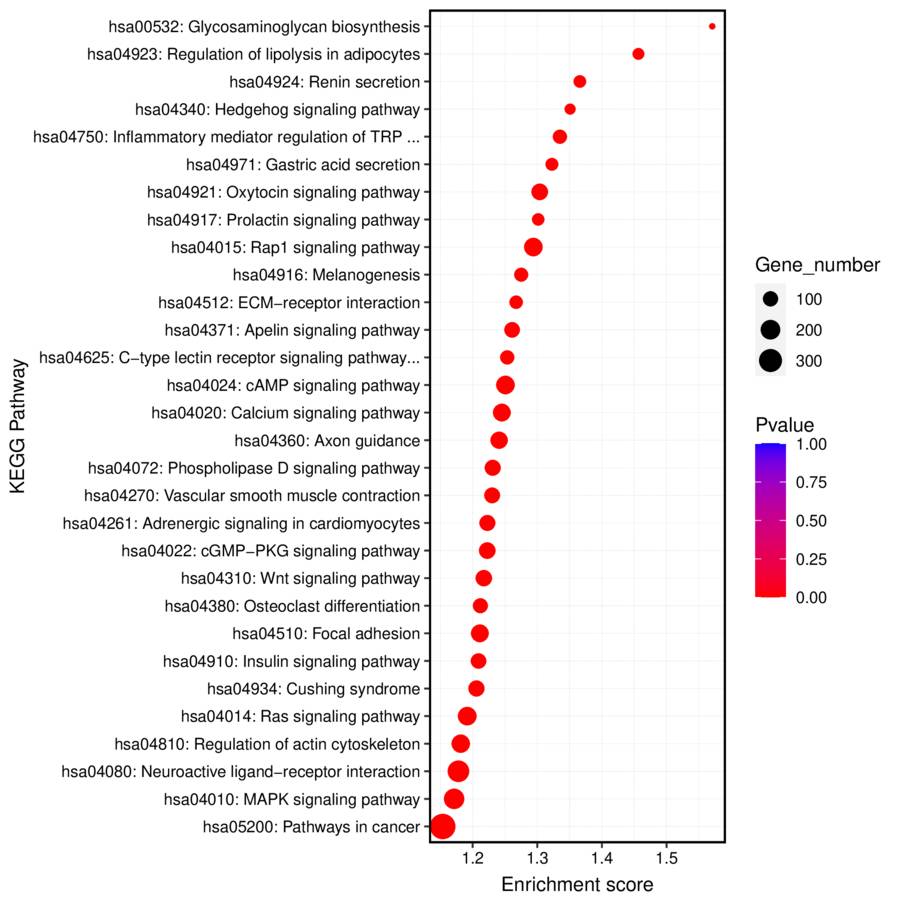 Fig3. KEEG Biological Pathway Enrichment Analysis of Differential Peak Related Genes.
Fig3. KEEG Biological Pathway Enrichment Analysis of Differential Peak Related Genes.
- Motif Analysis
The peak sequence of clip-seq was extracted, and the motif analysis was conducted using Homer software. The smaller the P value, the more significant of the motif. The meaning of motif sequence can be interpreted by reading the literature or queried using tomtom database.
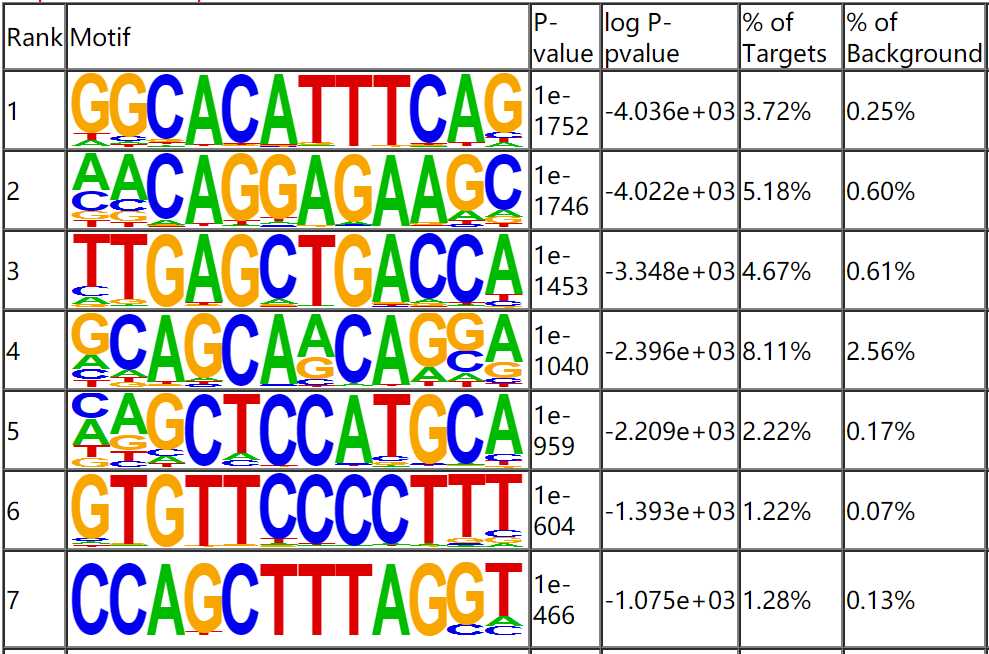 Fig4. Motif Analysis.
Fig4. Motif Analysis.
- Protein Binding RNA Analysis and Seq Result Visualization
The CHIRP-Seq analysis identified enriched RNA targets bound by the protein of interest, with results visualized using IGVtools for clear interpretation.
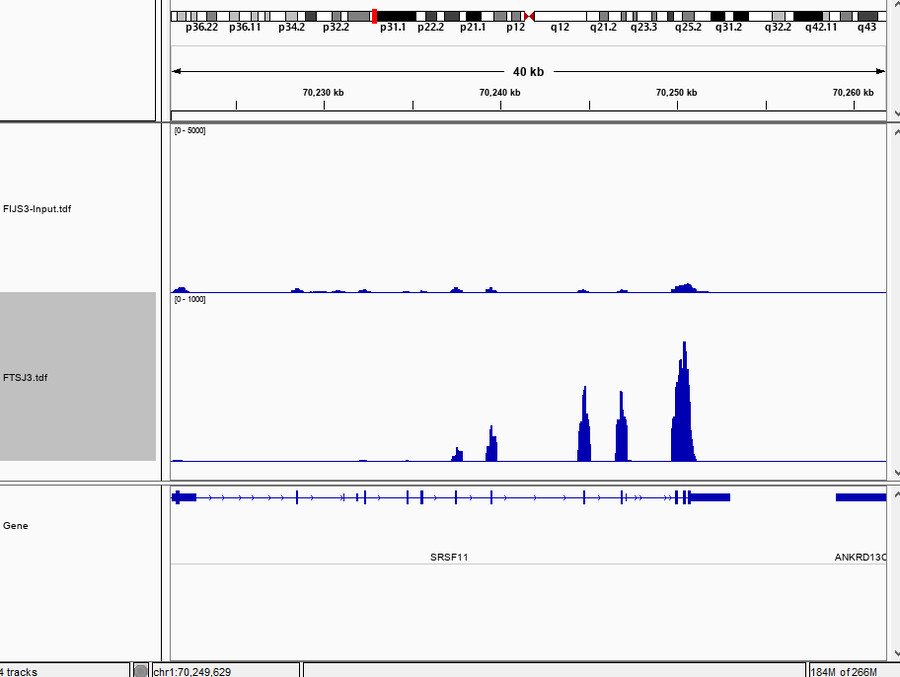 Fig5. Chirp-seq Result Visualization.
Fig5. Chirp-seq Result Visualization.
Conclusions and Discussions
In this project, we successfully performed ChIRP-Seq experiments to screen interaction DNAs of xxx lncRNA in xxx cells. The identified binding sites were analyzed through KEGG pathway and GO enrichment analyses, revealing potential biological functions. Motif analysis further identified conserved sequence motifs in the binding peaks, providing important clues for future research. These results collectively enhance our understanding of the regulatory roles of xxx lncRNA in gene expression.
Customer Testimonials
"Their ChIRP-Seq service uncovered novel lncRNA-chromatin interactions in our cancer models. The integrated analysis saved us months of work! Highly recommend for epigenomics projects."
--- Dr. Emily Zhang, Principal Scientist, Cancer Research Institute
"Flawless technical support and FAST turnaround. The high-resolution data validated our hypotheses about RNA-guided chromatin looping in neurodevelopment."
--- Prof. Raj Patel, Director, Biomedical Research Center
"Cost-effective and reliable. Their plant-optimized ChIRP-Seq protocol detected low-abundance RNA-DNA crosslinks in Arabidopsis-critical for our crop engineering program."
--- Dr. Lena Müller, Head of Genomics, BioPlant Labs
"Outstanding ROI! The team helped us map viral RNA integration sites in host genomes using ChIRP-Seq, accelerating our antiviral drug discovery pipeline."
--- Mr. James Carter, CTO, Virome Diagnostics
"Their expertise in peak annotation and regulatory element analysis transformed our cardiovascular epigenetics study. A+ for data accuracy and customer service."
--- Dr. Sofia Alvarez, Senior Researcher, CardioGenomics Institute
"We compared 3 vendors; their ChIRP-Seq had the lowest noise-to-signal ratio. The lncRNA mapping data is now central to our biomarker discovery platform."
--- Ms. Priya Kapoor, CSO, Pharma Innovations
FAQs
Resources
Reference:
- Chu C.; et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667-678.