Electrophoretic Mobility Shift Assay (EMSA) Service
Background
Introduction
Profacgen offers a handy Electrophoretic Mobility Shift Assay (EMSA) service, ideal for exploring those complex protein-nucleotide interactions. It's super useful if you're researching gene regulation, transcription factor binding, or the details of DNA repair and RNA processing. They handle everything from prepping your samples and labeling nucleotides to running the electrophoresis and breaking down the data. So, you can count on them for top-notch results that'll really drive your research forward.
What is Electrophoretic Mobility Shift Assay (EMSA)?
EMSA is a lab method that helps researchers spot and study how proteins interact with nucleic acids. Basically, you take a labeled nucleic acid probe, mix it with a protein sample, and then run it through a non-denaturing gel. If the protein binds to the probe, the combo moves slower through the gel than the free probe, making a clear band shift. You can see this shift using techniques like autoradiography, fluorescence, or chemiluminescence. This movement tells you there's a protein-nucleic acid interaction going on, offering clues about how specific and strong the binding is.
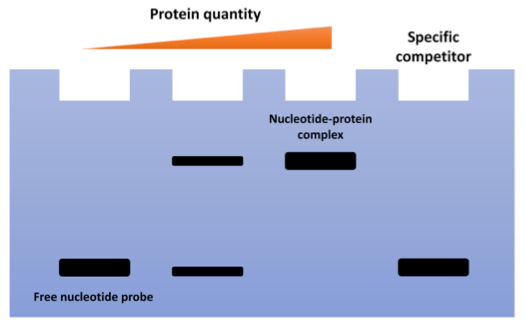 Fig1. Schematic Diagram of EMSA.
Fig1. Schematic Diagram of EMSA.
Applications of EMSA
EMSA is quite handy in molecular biology and biochemistry:
- Gene Regulation and Transcription Factor Binding: With EMSA, researchers can check out how transcription factors attach to specific DNA sequences, giving insights into gene regulation.
- RNA-Protein Interactions: It's handy for looking into RNA-protein interactions, which are crucial for grasping RNA processing and how stable it is.
- DNA Repair Mechanisms: EMSA helps reveal how DNA repair proteins hook onto damaged DNA sequences, providing a clearer picture of how DNA repair works.
- Drug Screening and Target Validation: It's also used to find compounds that change protein-nucleotide interactions, important for drug discovery and target checks.
- Cell Signaling Pathways: EMSA allows for the analysis of protein-protein interactions in cell signaling, helping us understand how cells communicate.
Service Procedure
Sample Requirements:
- Protein Sample: Purity >90%, concentration >0.3 mg/ml.
- Cell/Tissue Samples: Cell samples should include >107 cells; tissue samples should be >100 mg.
- Nucleotides: Nucleotides used as probes should be sub-cloned into plasmids and verified by DNA sequencing, or provide nucleotide information (gene name, source, GenBank accession number, sequence, etc.).
Deliverables:
- High-resolution EMSA data.
- Detailed experimental reports, including binding affinity and interaction quantification.
- Visualizations of electrophoretic bands and binding interactions.
Service Details:
| Nucleotide and Protein Preparation (Optional) | Quality Control Testing (EMSA will be stopped if QC fails) | Nucleotide Labeling and Purification | Electrophoresis of Nucleotide-Protein Complexes | Membrane Transfer and Cross-Link by UV (Optional) | Signal Detection | |
| Critical Points | Nucleotides and proteins should be stored at -20°C or -80°C to maintain stability. Avoid repeated freeze-thaw cycles to prevent degradation. | If QC fails, troubleshoot by checking sample preparation protocols, storage conditions, and reagent quality. | Ensure complete labeling and purification to avoid background noise in EMSA results. | Ensure proper mixing of reagents and avoid vigorous pipetting to prevent disruption of protein-nucleotide complexes. | Handle the membrane with clean forceps and powder-free gloves to avoid contamination. Ensure even UV exposure to prevent uneven cross-linking. | Follow the manufacturer's instructions for optimal detection conditions. Ensure that the membrane is not overexposed to prevent signal saturation. |
Why Choose Profacgen?
- State-of-the-Art Platform: Utilize advanced technological platforms for high-sensitivity and high-resolution results.
- Short Turnaround Time: Efficient workflow ensures quick delivery of results.
- Competitive Pricing: Best price in the market without compromising on quality.
- Consistent Results: Reliable and consistent results with detailed experimental reports.
- Customized Solutions: Tailored experimental schemes to meet specific research needs.
Case Study
Background
The aim of this project is to verify the interaction of DUX4 protein with its target DNA sequences using EMSA. The project involves designing and synthesizing a biotin-labeled oligo probe, incubating it with DUX4 protein, and detecting the resulting protein-DNA complex via chemiluminescence. Additionally, an unlabeled probe is used for competitive experiments to confirm specificity.
Results
- EMSA Results: The results showed distinct shift bands between DUX4 protein and the target probe. The shift bands were darker with increasing concentrations of DUX4 protein. Unlabeled probes at 50 nM, 0.5 μM, and 5 μM concentrations were used for competitive binding experiments, demonstrating partial to complete competition.
- Specific Binding: The results indicated that DUX4 protein specifically binds to the target sequence 5'-xxxxxxxxxxxxxxxx-3'.
Note: Key information has been blurred due to the confidentiality of the project promised to the client.
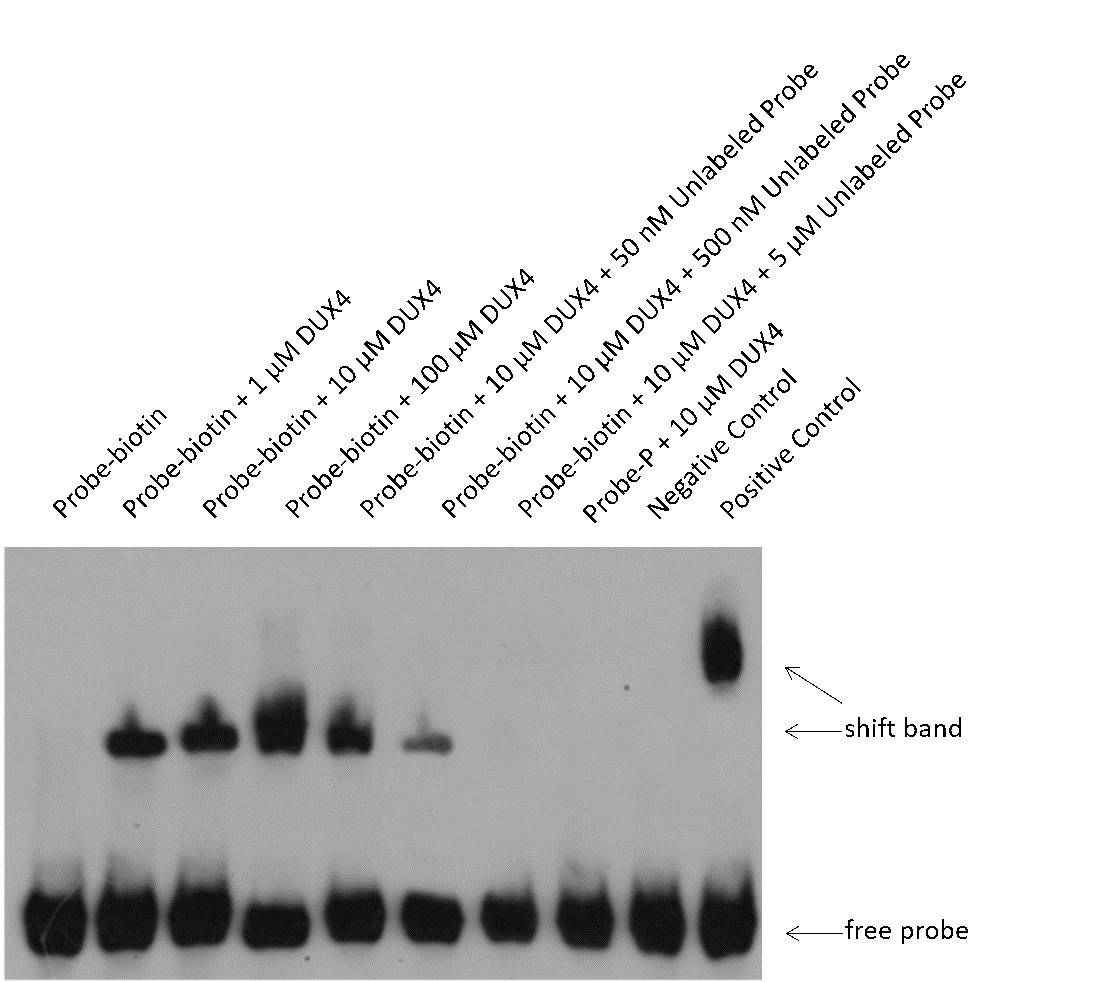 Fig2. EMSA Results for DUX4 Protein and Its Target Probe.
Fig2. EMSA Results for DUX4 Protein and Its Target Probe.
Background
The aim of this project is to verify the interaction of X protein with its target DNA sequences using EMSA. Specifically, four oligos were designed and synthesized in vitro, labeled with biotin at the 5'-end. After incubation with purified X protein, the resulting complexes were subjected to gel electrophoresis, and the shift band with DNA-X was detected using the chemiluminescence method. Additionally, unlabeled probes were used for competitive binding experiments.
Results
- Shift Band Detection: The results showed a distinct shift band in the sample containing probe2 and X protein, indicating a successful binding interaction. No shift bands were observed in the other samples (probe1, probe3, and probe4), suggesting a lack of interaction between these probes and X protein.
- Binding Specificity: The presence of the shift band with probe2 demonstrates that X protein specifically binds to the target sequence 5'-xxxxxxxxxxxxxxxxx-3'.
- Visual Representation: The EMSA results are illustrated in Figure 3, which shows the distinct shift band for the interaction between X protein and probe2.
Note: Key information has been blurred due to the confidentiality of the project promised to the client.
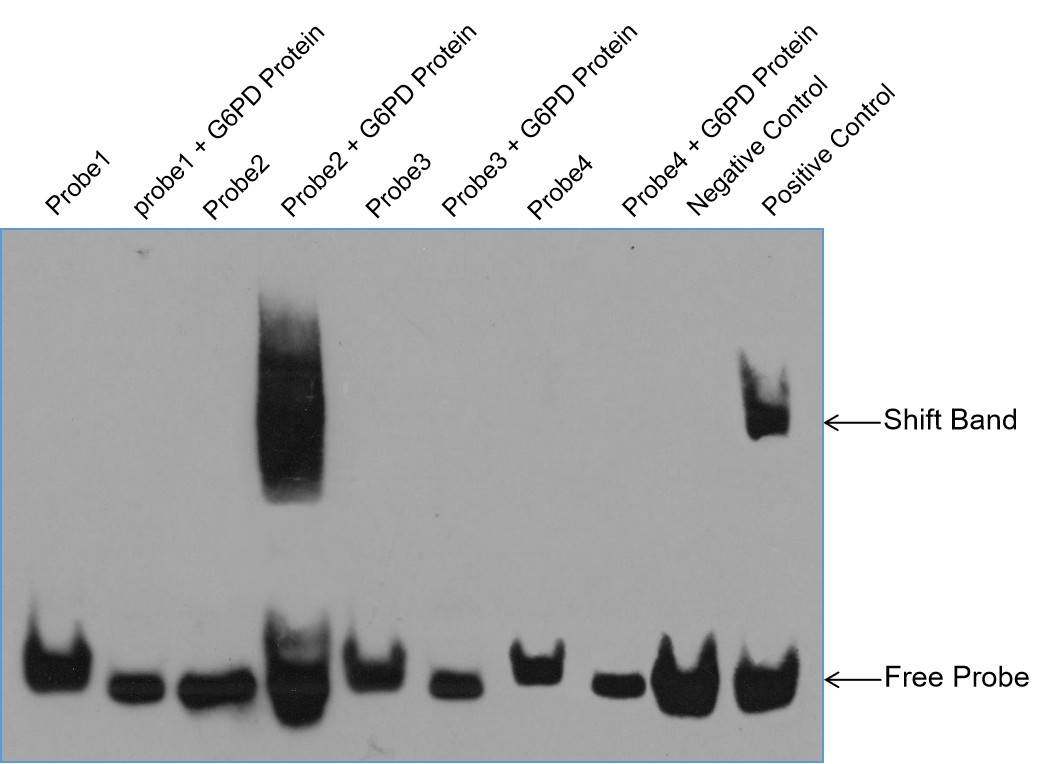 Fig3. EMSA Results for X Protein and 4 Probes.
Fig3. EMSA Results for X Protein and 4 Probes.
FAQs
Resources
| DNA/RNA pull down | DNase I Footprinting Assay Service | Chromatin Immunoprecipitation (ChIP) Assay Service | CUT&RUN-Sequencing Service | DAP-seq Service (DNA affinity purification sequencing) |
References:
- Mansouri-Noori F.; et al. Electrophoretic mobility shift assays (EMSAs) for in vitro detection of protein-nucleic acid interactions. STAR Protoc. 2024;5(2):103128.
- Stowell JAW.; et al. Gel-Based Analysis of Protein-Nucleic Acid Interactions. Methods Mol Biol. 2021;2263:321-339.
References from our customer:
Sirisena ND.; et al. Electrophoretic mobility shift assays implicate XRCC2: rs3218550C> T as a potential low-penetrant susceptibility allele for sporadic breast cancer. BMC Res Notes. 2019;12(1):476.
XRCC2 is known to play an essential role in homologous recombination repair of DNA double-strand breaks. It is plausible that this variant may be exerting regulatory effects on XRCC2 gene expression leading to altered DNA repair capacity. Further functional studies are warranted to validate this finding. In order to investigate whether a single nucleotide variant at the rs3218550 site may alter the protein-DNA interactions, EMSA was performed by Profacgen.