Proximity-dependent Biotin Identification (BioID) Service
Background
Introduction
In the rapidly evolving field of protein interaction research, Profacgen delivers cutting-edge Proximity-dependent Biotin Identification (BioID) services to uncover intricate cellular networks and dynamic molecular interactions. Leveraging advanced BirA* enzyme systems and streamlined workflows, we empower researchers to map protein interactomes with high specificity, even in challenging biological models. Our expertise spans diverse applications, from target validation to pathway mechanism studies, ensuring actionable insights for drug discovery and functional genomics.
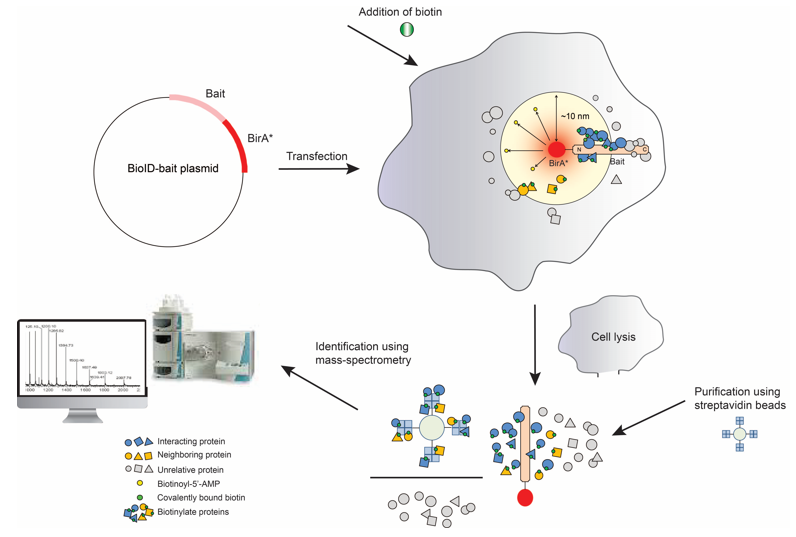
Technical Workflow of BioID
- Phase 1: Fusion Protein Construction
BioID begins by genetically fusing a promiscuous biotin ligase (BirA*, an R118G mutant of E. coli BirA) to a target protein. This fusion ensures BirA* localizes to the target's native cellular compartment.
- Phase 2: Proximity Biotinylation
In live cells, BirA* uses ATP and exogenous biotin to generate reactive biotin-5′-AMP, which labels lysine residues on proteins within a ~10–20 nm radius. The short-lived intermediate ensures spatial specificity.
- Phase 3: Protein Capture
Cells are lysed, and biotinylated proteins are isolated using streptavidin-coated beads. Harsh washes remove nonspecific interactions, enriching the target's proximal interactors.
- Phase 4: Mass Spectrometry Analysis
Purified proteins are digested into peptides, analyzed by LC-MS/MS, and identified via database matching. Statistical filtering distinguishes true interactors from background.
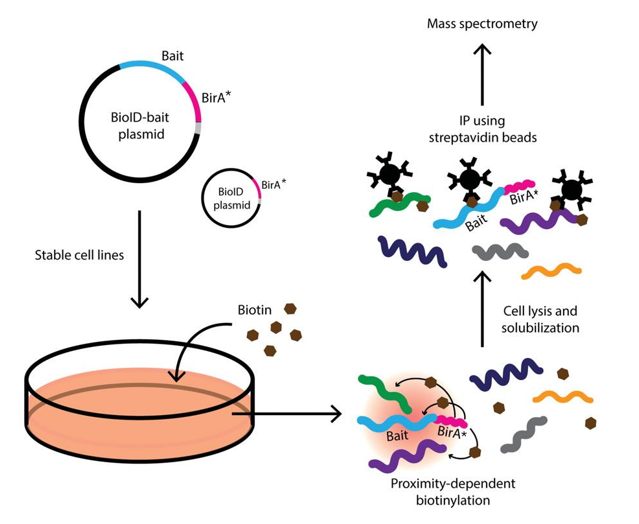 Fig2. Diagram of Basic Protocol. (Le Sage, et al., 2016)
Fig2. Diagram of Basic Protocol. (Le Sage, et al., 2016)
Advantages of Profacgen's BioID Service
1) In Vivo Interaction Mapping: Detect protein interactions in living cells, reflecting true biological conditions.
2) High Sensitivity and Specificity: Identify low-abundance proteins with minimal false positives/negatives.
3) Versatility Across Species: Suitable for a wide range of organisms and cell types, including mammalian cells, plants, and yeast.
4) Comprehensive Support: Offer complementary validation services (e.g., IP assays) to confirm interactions and provide actionable insights.
Service Procedure
Sample Submission Requirements
To activate the BioID service, clients must submit the following materials:
Plasmid Containing Target Gene
- Provide a plasmid carrying the gene of interest designed for fusion with the BirA* tag.
- Confirm that the plasmid sequence is validated (e.g., via Sanger sequencing) to ensure absence of unintended mutations or sequence errors.
Cell Line Information
- Specify the cell line for stable fusion protein expression (e.g., HEK293, HeLa, HepG2).
- For custom or engineered cell lines, provide detailed handling protocols, including culture conditions, transfection methods, or selection markers (if applicable).
Additional Reagents (Optional)
If specialized reagents (e.g., antibodies for Western blotting) or detection tools (e.g., fluorescent probes for imaging) are required for post-analysis, either supply these materials or explicitly list their specifications (e.g., product catalog numbers, dilution ratios).
Note: If clients cannot provide specific reagents, we can prepare a full set.
Delivery
- Project Report: Comprehensive report detailing the experiment's background, goals, methods, results, and conclusions.
- Silver-Stained Gel Images: Visual documentation of silver staining results to confirm biotinylation and protein capture.
- Mass Spectrometry Data: Raw MS data files and detailed analysis results for protein identification.
- PPI Network Diagrams: Graphical representation of protein-protein interactions based on MS findings.
- Stable Cell Line Validation: Fluorescence microscopy and Western Blot data confirming target protein expression in stable cell lines.
- Experimental Protocols: Step-by-step methods and reagent details for reference and reproducibility.
- Supplementary Analysis Files: Additional tables and data to support in-depth interpretation of results.
Service Scope
| Vector Construction |
|
| Stable Cell Line Development |
|
| Biotinylation & Protein Capture |
|
| Mass Spectrometry Analysis |
|
| Data Interpretation |
|
Why Choose Profacgen?
Technical Expertise in Proximity-Dependent Labeling
Profacgen's team specializes in BioID technology, with validated protocols for optimizing BirA* fusion constructs, biotinylation kinetics, and background reduction, ensuring precise identification of proximal interactors.
End-to-End Customization
From vector design (e.g., choice of promoters, tags) to cell line selection (primary, cancer, or engineered lines), workflows are tailored to match experimental needs, including niche applications like organelle-specific interactome mapping.
Rigorous Multi-Stage QC
Every step undergoes stringent validation: Sanger sequencing for plasmid integrity, Western blot for fusion protein expression, and label-free quantitation (LFQ) in MS to filter low-confidence hits, minimizing false positives.
Advanced Proteomics Infrastructure
Equipped with state-of-the-art Orbitrap mass spectrometers and AI-driven bioinformatics pipelines, we deliver high-depth proteomic coverage and statistically robust interaction networks (SAINT, CRAPome-filtered).
Turnkey Service with Timely Delivery
Fixed turnaround times (e.g., 8-10 weeks for full workflow), dedicated project managers, and interactive reporting portals ensure transparency and accelerate downstream validation studies.
Case Study
* NOTE: We prioritize confidentiality in our services to safeguard technology and intellectual property for enhanced future value and protection. The following case study has been shared with the client's consent.
Goal
The primary objective of this project was to identify interaction proteins of the target protein (xxx) in xxx cells using the BioID (Proximity-dependent Biotin Identification) technique. The process involved fusing the target protein with a biotin ligase (BioID), constructing stable cell lines using lentivirus, and identifying biotinylated proteins via mass spectrometry.
Results
- Silver Staining
Two rounds of silver staining were performed to visualize the biotinylated proteins captured by streptavidin beads. The results demonstrated successful biotinylation and enrichment of target proteins.

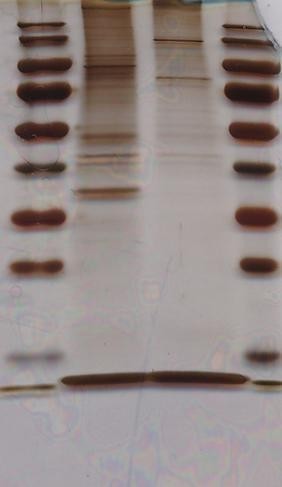
Fig3. Two Siliver Stainning Results.
- Mass Spectrometry
Mass spectrometry analysis identified a comprehensive list of proteins interacting with the target protein. Detailed results, including protein identification and analysis instructions, are provided in the undisclosed folder.
Conclusions and Discussions
The project successfully constructed stable cell lines expressing the target protein fused with BioID. The biotinylation and enrichment process effectively captured interacting proteins, which were subsequently identified by mass spectrometry. The results provide valuable insights into the protein-protein interactions of the target protein in xxx cells, achieving the project's goals.
Goal
The primary goal of this project was to identify the interacting proteins of the target protein (xxx) using the BioID technique. This involved constructing a stable cell line expressing the target protein fused with a biotin ligase (BirA*), followed by biotinylation, protein capture, and identification via mass spectrometry.
Results
- Vector Construction
The target gene (xxx) was successfully cloned into the vector H6431 pLenti-EF1a-EGFP-F2A-Puro-CMV-MCS-BirA-HA. The construct was verified through sequencing and vector mapping. - Stable Cell Line Construction
Stable cell lines (H28178 and H6431) were established using lentivirus-mediated transduction.
Infection efficiency reached 80% under MOI 20 conditions, and stable expression was confirmed by fluorescence microscopy and Western Blot analysis.
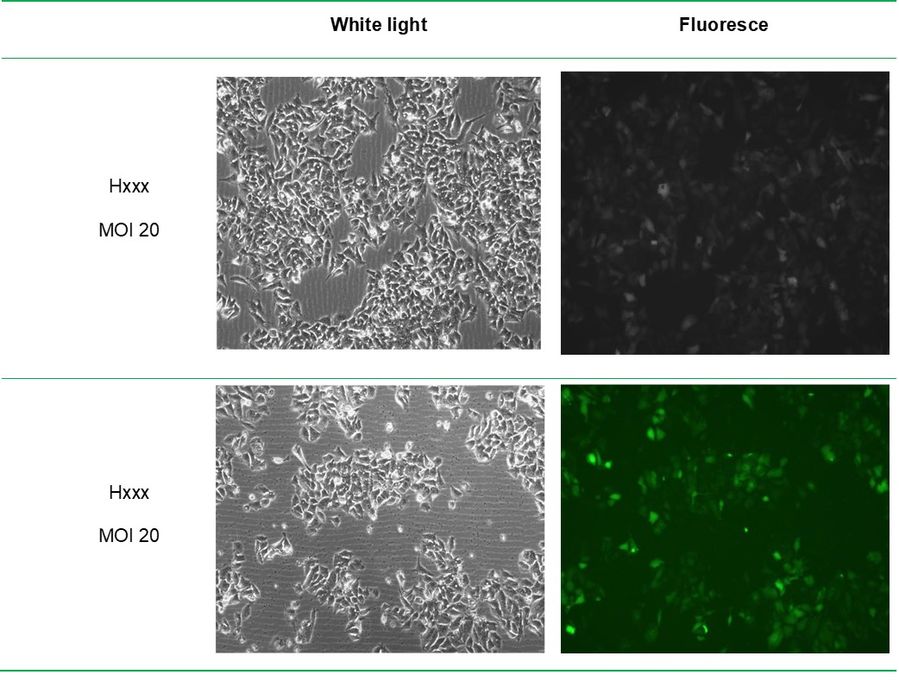 Fig4. Lentivirus infection of xxx cells for 72h.
Fig4. Lentivirus infection of xxx cells for 72h.
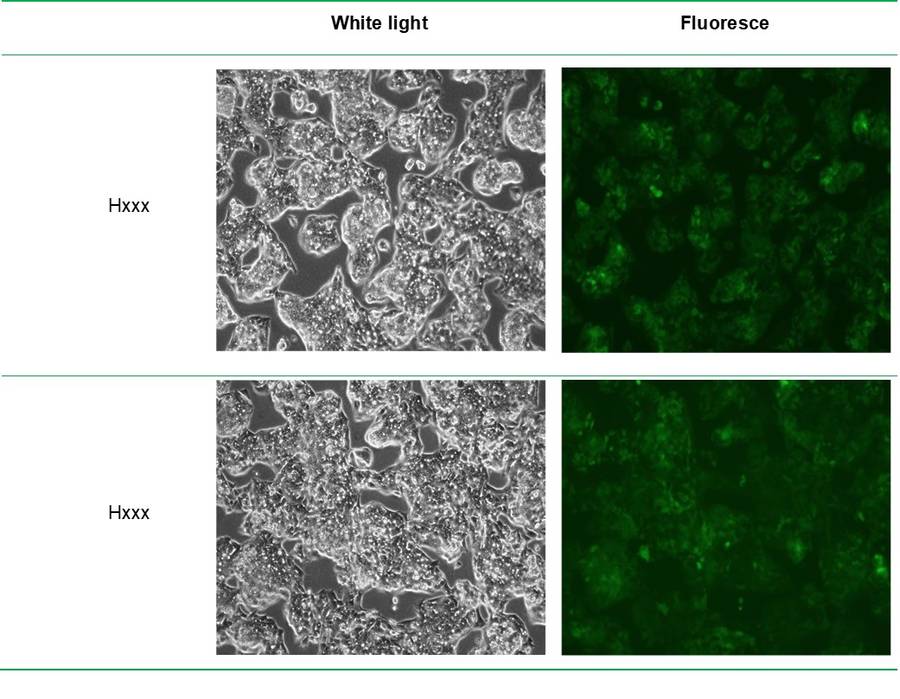 Fig5. Fluorescence photos of stable strains after 14 days of screening.
Fig5. Fluorescence photos of stable strains after 14 days of screening.
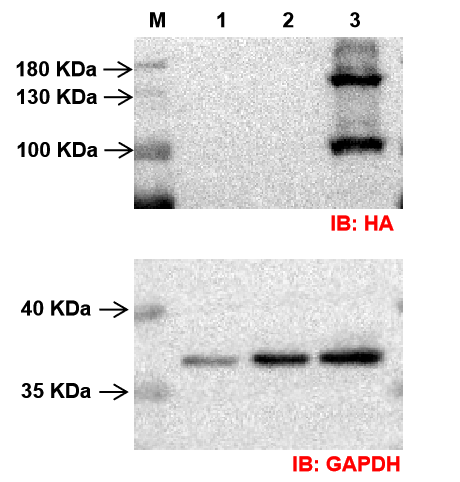 Fig6. The results of the Western Blot assay.
Fig6. The results of the Western Blot assay.
- BioID Analysis
Biotinylation was performed in stable cell lines, followed by protein capture using streptavidin beads.
Silver staining of SDS-PAGE gels confirmed the presence of biotinylated proteins. - Mass Spectrometry Identification
Mass spectrometry identified a comprehensive list of interacting proteins from the target cell line (H28178) and control cell line (H6431).
Differential analysis highlighted specific proteins interacting with the target protein (xxx).
Conclusions and Discussions
The project successfully achieved its goal of identifying interacting proteins of the target protein (xxx) using the BioID technique. The constructed stable cell lines and validated constructs provided a reliable platform for biotinylation and subsequent protein identification. Mass spectrometry analysis yielded valuable insights into the protein-protein interactions, demonstrating the effectiveness of the BioID approach in capturing transient and weak interactions in a native cellular environment. The results lay the groundwork for further studies into the functional roles of these interactions in cellular processes.
Customer Testimonials
"Profacgen's BioID service provided exceptional clarity in mapping immune cell protein interactions. Their ability to handle primary cell models with high background complexity was impressive. The data analysis included annotated pathways directly relevant to our hypothesis, and their team accommodated last-minute adjustments to the experimental design. A trusted resource for rigorous academic collaboration."
Dr. Laura Bennett | Principal Scientist, Academic Immunology Center
"We needed precise interactome data to validate a drug target's mechanism. Profacgen's BioID workflow delivered reproducible results with minimal non-specific binding, even for membrane-associated proteins. Their cross-validation with orthogonal methods strengthened our confidence in the findings. Timely communication and adherence to ethical data-sharing protocols were additional strengths."
Prof. Hiroshi Tanaka | Director, Molecular Pharmacology Lab (University Hospital)
"Speed and cost transparency were critical for our seed-funded team. Profacgen's BioID service identified key interactors in under 10 weeks, accelerating our preclinical pipeline. Their tiered pricing model allowed us to scale experiments as funding permitted. The team also provided actionable advice on optimizing bait protein expression-something rarely offered at this price point."
Dr. Fatima Al-Mansoori | Co-Founder, Precision Medicine Startup
"Profacgen adapted BioID for pathogen-host interaction studies, a niche application requiring biosafety-level containment. Their optimized protocol reduced false positives compared to our in-house attempts. Raw mass spectrometry data was delivered in standardized formats, easing integration with our bioinformatics tools. A collaborative partner for high-containment research."
Dr. Nathan Carter | Lead Investigator, Infectious Disease Research Institute
"We outsource specialized assays to maintain agility. Profacgen's BioID service consistently met our clients' demands for industry-grade data.. Their audit-ready documentation and GLP-compliant reporting align perfectly with pharmaceutical standards. The ability to process cryopreserved samples expanded our service offerings."
Ms. Elena Rossi | Head of Omics Services, Pharma CRO
"Profacgen's expertise in engineering protein proximity networks supported our enzyme optimization pipeline. Their customized BioID approach resolved spatial interactions in microbial chassis organisms, which traditional methods failed to capture. Budget-friendly without compromising on statistical rigor-exemplary for industrial R&D."
Dr. Kwame Osei | Senior Biologist, Industrial Biotechnology Company
FAQs
- Bait expression: Stable expression of the BirA*-fusion bait protein is required. Provide constructs with validated expression levels (e.g., via Western blot or fluorescence tagging).
- Cell viability: Maintain >85% viability during biotinylation (typically 18–24 hours). Avoid prolonged biotin treatment to minimize cytotoxicity.
- Biotin supplementation: Include biotin (e.g., 50 μM) in the culture medium during the biotinylation window.
For non-mammalian systems (e.g., plant cells or microbes), consult our team for protocol adjustments, including biotin concentration or incubation time optimization.
- Bioinformatic interpretation: Customized pathway enrichment analysis (GO, KEGG, Reactome) and network visualization tailored to your biological context.
- Hit prioritization: Guidance on filtering interactors (e.g., excluding common contaminants, focusing on novel/uncharacterized proteins).
- Experimental follow-up: Recommendations for orthogonal validation or functional assays to confirm relevance.
Additional support, such as cross-project meta-analysis or integration with public datasets, is available upon request.
- Negative controls: Cells expressing untagged BirA* or non-biotinylated bait.
- Background subtraction: Exclusion of proteins identified in controls or public contaminant databases (e.g., CRAPome).
- Statistical filtering: SAINTexpress or CompPASS algorithms to assign interaction confidence scores.
Resources
| Co-Immunoprecipitation (Co-IP) | Yeast Two-Hybrid Screening | Affinity- Purification /mass Spectrometry (AP-MS) Service | Protein Aggregation Analysis | Plasma Protein Binding Assay |
Reference:
- Le Sage V.; et al. Proximity-Dependent Biotinylation for Identification of Interacting Proteins. Curr Protoc Cell Biol. 2016;73:17.19.1-17.19.12.