The two-hybrid system is a genetic method that uses transcriptional activity as a measure of protein-protein interaction [1]. It relies on the modular nature of site-specific transcriptional activators, which consist of a DNA-binding domain and a transcriptional activation domain. The DNA-binding domain serves to target the activator to the promoter region of specific genes, and the activation domain contacts other proteins of the transcriptional machinery to enable transcription to occur. The two-hybrid system is based on the observation that the two domains of the activator do not have to be covalently linked and can be brought together by the interaction of any two interacting proteins.
The application of this system requires two hybrids to be constructed (see below): a DNA-binding domain fused to one protein, X, and a transcription activation domain fused to another protein, Y. These two hybrids are expressed in a cell containing one or more reporter genes. If the X and Y proteins interact, they create a functional activator by bringing the activation domain into close proximity to the DNA-binding domain, that can be detected by expression of the reporter genes. While the assay has been generally performed in yeast cells, it works similarly in mammalian cells.
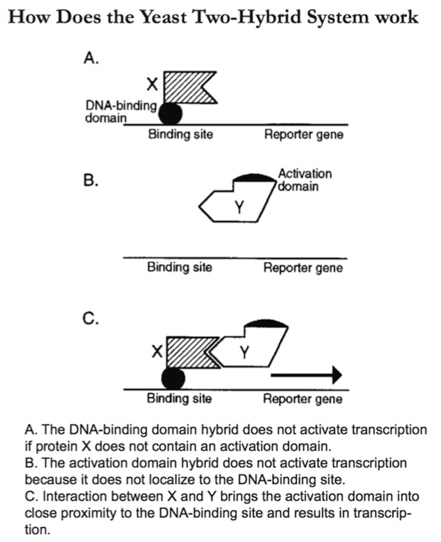
Two-hybrid systems have been used to probe the function of new proteins ever since they were developed [1,2].
This method can be used to detect interactions between candidate proteins whose genes are available by constructing the appropriate hybrids and to test reporter gene activity. If an interaction is detected, deletions can be made in the DNA encoding one of the interacting proteins to identify a minimal domain for interaction. In addition, point mutations can be made to identify specific amino acid residues critical for the interaction. Most significantly, the two-hybrid system can be used to screen libraries of activation domain hybrids to identify proteins that bind to a protein of interest. These screens result in the immediate availability of the cloned gene for any new protein identified.
The two-hybrid system has several features that make it useful for analysis of protein-protein interactions [3]. It is highly sensitive, capable of detecting interactions that are not detected by other methods. The use of protein fusions also means that the site of interaction may be occluded by one of the transcription factor domains. Interactions depending on a posttranslational modification that does not occur in yeast cells will not be detected, though. Sometimes, proteins, including those not normally involved in transcription, will activate transcription when fused to a DNA-binding domain, and this activation prevents a library screen from being performed. However, it is often possible to delete a small region of a protein that activates transcription while retaining other properties of the protein.
Most commonly used versions of the two-hybrid system all involve DNA-binding domains that derive from the yeast Gal4 protein or the E. coli LexA protein. Transcriptional activation domains are commonly derived from the Gal4 protein or the herpes simplex virus VP16 protein. Reporter genes include the E. coli lacZ gene and selectable yeast genes such as HIS3 and LEU2. An increasing number of activation domain libraries are now feasible for proteins from many different organisms or specific mammalian tissues.
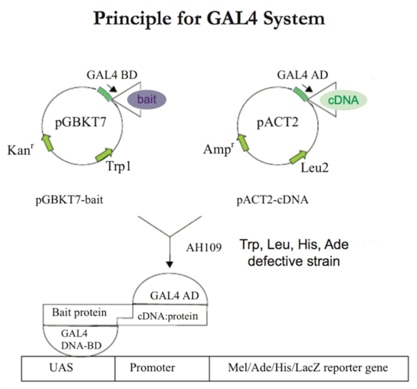
The following protocol can be a reference for you to know more about the technical route for traditional yeast two-hybrid screening.
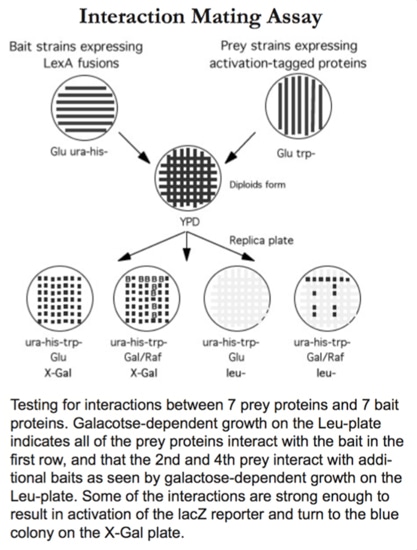
1. Interaction mating assay- small numbers of bait or prey proteins.
Refer to the following for the testing of a small set of proteins for interactions. The test set may include different allelic forms of the original bait, a set of structurally related proteins, a set of proteins known or suspected to be involved in a biological process, or unrelated proteins used to demonstrate the specificity of an interaction.
| 1) | Streak different bait strains in horizontal parallel stripes on a plate lacking uracil and histidine (-u-h Glu). Create a duplicate plate of bait strains for each different plate of prey strains to be used. |
| 2) | Streak different prey strains in vertical parallel stripes on a plate lacking tryptophan (-w Glu). |
| 3) | Include a prey strain that contains the prey vector encoding only the activation domain. |
| 4) | Create a duplicate plate of prey strains for each plate of bait strains to be used. |
| 5) | Incubate the plates until growth is observed on the streaks. |
| 6) | Press a plate of prey strains to a replica velvet, ensure yeast from each streak are left on the velvet. |
| 7) | Press a plate of bait strains to the same replica velvet. |
| 8) | Lift the impression of the bait and prey strains from the velvet by pressing a YPD plate on it. Incubate for growth. |
| 9) | Replicate YPD plates to the diploid selection, indicator plates: -u-h-w Glu X-Gal, -u-h-w Gal/Raf. Ensure the YPD plate contains sufficient growth to enable a single impression on the velvet to be lifted by at least four indicator plates. |
| 10) | Patch control strains onto the indicator plates and incubate for growth. |
| 11) | Blue color will develop within 2 days when diploids grow. |
2. Interaction mating assay- large scale for hundreds of different bait or prey strains.
| 1) | Place large numbers of bait strains in a 96-well configuration. |
| 2) | Use the 96-prong device, sterilized in ethanol and flame, to transfer bait strains from the culture to the center of –u-h Glu plate. Each plate can contain 96 different bait strains. |
| 3) | Incubate the plates until all bait strains have grown to colonies with diameter close to 5 mm. Note that several positions on each plate should contain control strains with baits that activate various levels of transcription. |
| 4) | Inoculate a prey strain in –w Glu liquid media and grow with shaking. |
| 5) | Pour the culture into plate and then transfer to –w Glu plates. |
| 6) | Combine the bait and prey strains to a YPD plate. |
| 7) | Allow growth till diploid turns out by X-Gal indicator. |
Screening a protein against a panel of bait strains enables one to quickly test its ability to interact with a large number of know proteins. While the number of proteins in the existing panel is far less than the number of proteins in a cDNA library, the approach does offer the advantage of screening the test protein against a set of proteins enriched for those of current interest to the biological community. Now it is possible to identify interacting partners that have not yet been isolated from the same species, or that are not expressed in tissues from which interaction libraries have been made.
Please also note that: Since Gal4 system utilizes Gal4 as the DNA binding domain, it cannot be used with yeast strains that have a wild-type GAL4 gene. Also, since GAL4 is required to activate the GAL1 promoter, it cannot work for systems that use the GAL1 promoter to drive expression of the prey protein.
3. Interpreting interaction data.
Interaction between bait and prey results in the interaction phenotypes: growth of the strain on medium lacking leucine, and transcriptional activation of the lacZ reporter and production of active β-galactosidase.
The two-hybrid approach can produce false positive results, though, interactions that do not occur in biological settings. This should not be regarded as a significant drawback of genome-wide two-hybrid approaches, provided that the information in a protein linkage map may provide clues for important interactions which can be explored and further confirmed by other methods.
Click here to contact us for more technical information.
[1] Fields S, Song O. A novel genetic system to detect protein-protein interactions[J]. Nature, 1989, 340(6230): 245-6.
[2] Chien C T, Bartel P L, Sternglanz R, et al. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest[J]. Proc Natl Acad Sci U S A, 1991, 88(21): 9578-82.
[3] Kolonin M G, Zhong J, Finley R L. Interaction mating methods in two-hybrid systems[J]. Methods Enzymol, 2000, 328: 26-46.
Fill out this form and one of our experts will respond to you within one business day.