Post-transcriptional regulation is really important for gene expression and impacts development, differentiation, and a cell's response to environmental cues. It's a team effort involving RNA-binding proteins (RBPs), mRNA, and non-coding RNAs like microRNAs. These players all affect RNA processes like splicing, how RNA exits the nucleus, where it ends up in the cell, mRNA decay, and translation. Traditional methods to study these interactions have their limitations, highlighting the need for more advanced techniques. RIP-Seq (RNA Immunoprecipitation Sequencing) is a game-changer here. It helps researchers find and analyze RNA targets of RBPs directly in living cells. Profacgen leads in the area of protein-nucleic acid interactions and offers a top-notch RIP-Seq service, helping researchers dig deeper into the complex world of post-transcriptional regulation.
RIP-Seq is a high-tech, in vivo method for finding specific RNAs linked to RNA-binding proteins (RBPs). It works by using antibodies to pull out mRNP complexes that have naturally formed between proteins and RNAs. This step gathers up RNA molecules attached to the RBPs, making it easier to clean up and analyze these RNA targets. Next, these RNA pieces are turned into cDNA and run through next-generation sequencing, giving researchers a big-picture look at RNA-protein interactions. Compared to older techniques, RIP-Seq is great because it finds RNA targets right in their natural environment, processes a lot of data quickly, and works with different RNA types like mRNA and non-coding RNAs. By using RIP-Seq, scientists can get a better understanding of how genes are controlled after they're transcribed and how they're expressed.
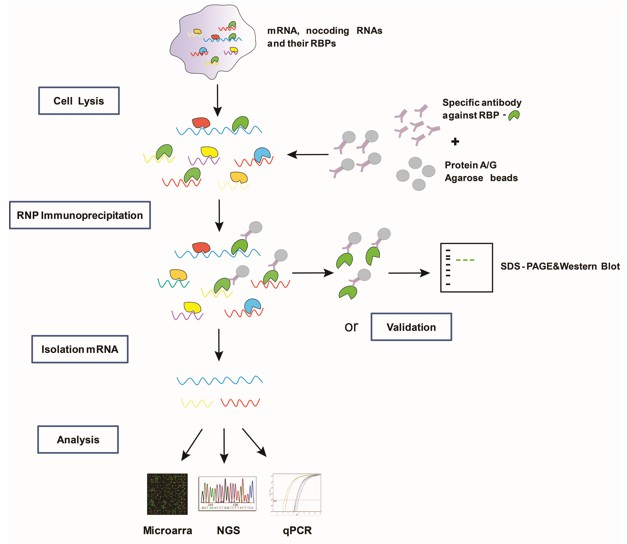 Fig1. The overall scheme of RIP-ChIP/Seq steps in detail.
Fig1. The overall scheme of RIP-ChIP/Seq steps in detail.
Quick RNA Target Discovery
RIP-Seq allows you to identify specific RNA targets associated with RNA-binding proteins (RBPs) quickly, giving you a clear overview of the RNAs connected to a particular RBP.
Easier Experimental Setup
One of the nice things about RIP-Seq is that it doesn't need UV cross-linking. This makes the whole process easier and more straightforward, which helps reduce complications and boost the chances of success.
Reliable and Sensitive
RIP-Seq produces high-quality data with a strong signal-to-noise ratio, making it easier to accurately pinpoint real RNA-protein interactions. This ensures that the results are trustworthy and can be repeated.
Flexible Across Research Areas
You can use RIP-Seq in various research fields, whether you're looking into post-transcriptional regulation, identifying miRNA targets, or examining how RBPs interact with non-coding RNAs.
In-Depth Data Insights
RIP-Seq generates detailed data that you can analyze to find RNA targets, perform peak calling, and run functional enrichment analyses. This helps deepen our understanding of the biological significance of RNA-protein interactions.
Figuring Out RNA Targets
RIP-Seq is like a detective tool for spotting the RNA buddies of RNA-binding proteins, whether they're mRNA or non-coding RNAs. This helps us see which RNAs are under the watch of certain RBPs.
Understanding Post-Transcriptional Moves
This method sheds light on what happens after transcription. It shows how RBPs mess with stuff like mRNA stability, where it goes, and how it's translated.
Exploring Non-Coding RNAs
RIP-Seq helps us find out how RBPs and non-coding RNAs, like microRNAs, hang out together, which is super important for gene regulation.
Checking Out What RBPs Do
By spotting which RNAs are linked with RBPs, RIP-Seq helps us understand what these proteins are up to in different biological gigs, like how they help with development and differentiation.
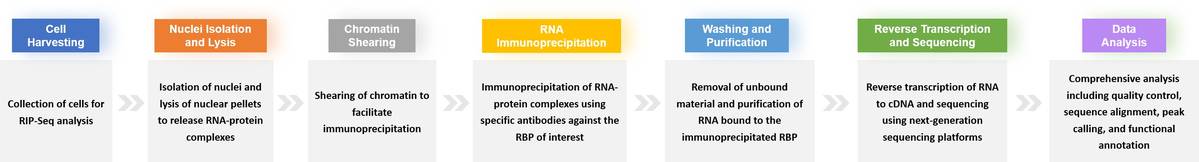
High-quality sequencing data.
Detailed reports on RNA-protein interactions.
Functional annotations and pathway analysis.
Comprehensive Data Analysis Services
We offer RIP-Seq data analysis services that include differential expression and pathway enrichment analysis. Additionally, we can customize our approach to fit your unique requirements, whether you need help identifying RNA targets or exploring RNA-protein interactions.
Combination with Other Omics Technologies
We integrate RIP-Seq with other omics technologies like proteomics and metabolomics, offering a fuller picture of cellular processes. Our joint analysis services combine RIP-Seq data with other omics data to reveal complex regulatory networks.
Background
The aim of this project is to screen for interaction RNAs, such as mRNA of xxx protein in target cells, using RIP-seq. Briefly, the RNA-xxx complex was separated by xxx antibody, and the bound RNAs were analyzed by high-throughput sequencing.
Results
The sequencing data was aligned to the reference genome, and the coverage area and depth were evaluated.
The number of reads in RIP-Seq and Input samples was obtained, and enriched target RNA was identified.
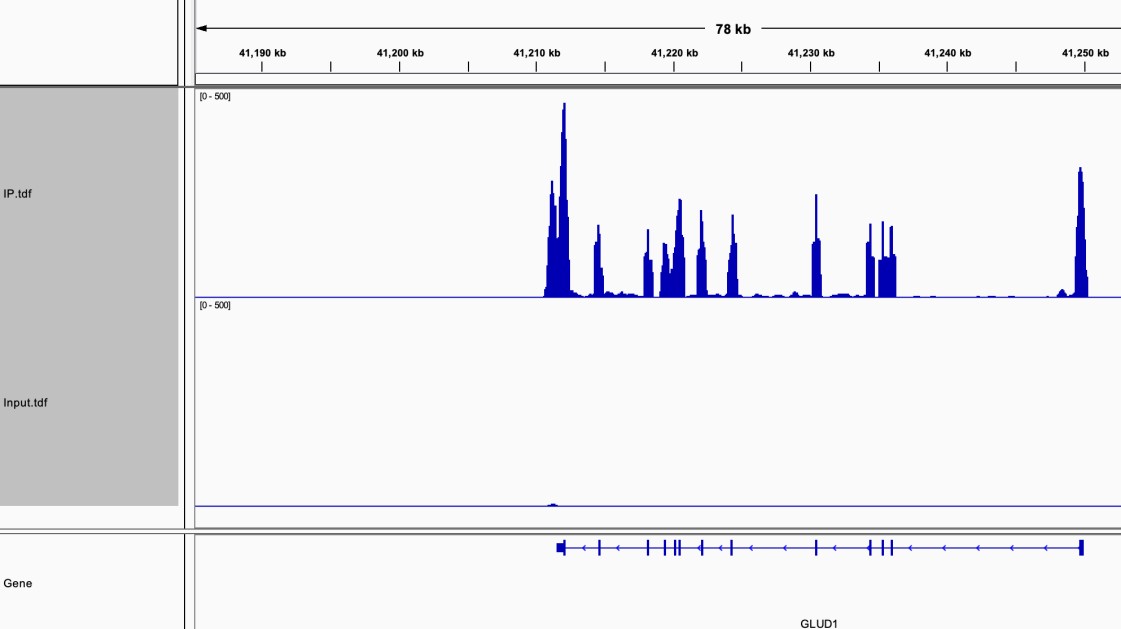 Fig2. The RIP-seq results were visualized using IGVtools.
Fig2. The RIP-seq results were visualized using IGVtools.
KEGG pathway enrichment analysis was conducted, and the most related biological pathways were identified.
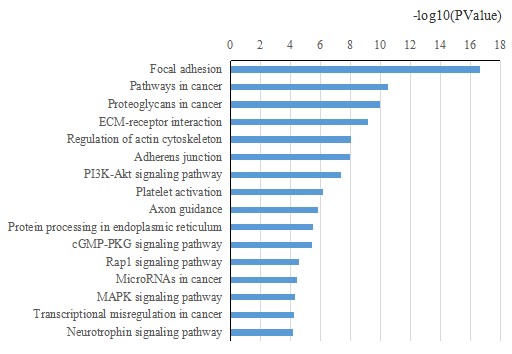 Fig3. KEEG Biological Pathway Enrichment Analysis.
Fig3. KEEG Biological Pathway Enrichment Analysis.
GO enrichment analysis was performed, and the most significant GO terms were identified.
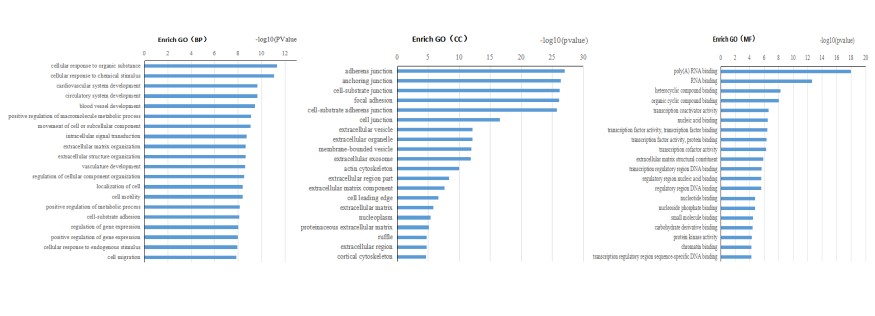 Fig4. GO Enrichment Analysis of Differential Genes.
Fig4. GO Enrichment Analysis of Differential Genes.
References:
Fill out this form and one of our experts will respond to you within one business day.