Nucleus-based Yeast Two-Hybrid Screening
Background
Introduction
Protein interaction analysis remains a cornerstone of functional genomics, with the nucleus-based yeast two-hybrid (Y2H) system standing as a pivotal tool for detecting direct molecular interactions. Originating from the groundbreaking work of Stanley Fields and Ok-Kyu Song in the early 1990s, this method has evolved into a high-throughput screening platform, enabling systematic mapping of interactomes under native nuclear conditions. By coupling bait-prey fusion constructs with selectable reporters, the system efficiently identifies binding partners while minimizing cytoplasmic interference, making it indispensable for unraveling signaling networks and drug target discovery.
Ready to explore protein networks with precision? Partner with Profacgen to leverage our expertise in high-throughput yeast two-hybrid screening and advanced functional genomics.
Unlock Your Project's Potential with Profacgen
Why Choose Nucleus-based Yeast Two-Hybrid Screening?
This method combines cost-effective screening with high sensitivity to detect weak/transient eukaryotic protein interactions in their native nuclear context. Customizable assays (e.g., promoter-reporter systems, PTM-compatible strains) ensure adaptability to diverse targets, while nuclear localization preserves physiologically relevant interactions, including post-translationally modified proteins. Ideal for mapping signaling hubs or drug targets, it bridges biochemical relevance with scalability unmatched by in vitro approaches.
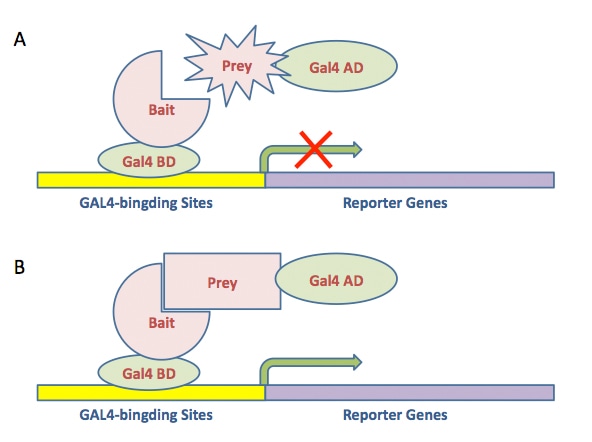 Fig1. GAL4 system was used to detect protein interaction in yeast two-hybrid experiment.
Fig1. GAL4 system was used to detect protein interaction in yeast two-hybrid experiment.
Applications Across Industries
| Types | Description |
| Biomedical Research |
|
| Agricultural Sciences |
|
| Industrial Biotechnology |
|
Service Procedure
Sample Submission Requirements
We're eager to assist you in exploring protein interactions with our Nucleus-based Yeast Two-Hybrid Screening service! To ensure a seamless start to your project, please provide the following information and samples:
- Target Protein Information
Kindly supply the gene sequence of your target protein, including the cDNA sequence. If you have any background information or specific requirements, please share them with us. This will enable us to customize our service to better meet your needs. - Bait Vector (Optional)
If you have already constructed a bait vector, please send it to us. If not, no worries-we can construct it for you! Just provide the necessary gene fragment details for primer design. - cDNA Library Selection
We offer a variety of standardized cDNA libraries for you to choose from. If you have a custom library, please let us know—we're happy to accommodate! - Sample Quality
For DNA samples, ensure they are high-purity with a concentration of ≥50 ng/µL. If submitting protein samples, make sure they are active and pure. If you have any questions, feel free to reach out—we're here to assist! - Additional Details
Any extra information about your target protein, such as known interactions or specific screening conditions, will be very helpful for us to optimize the process.
Delivery
- Detailed Screening Report
- Positive Clone Sequences & BLAST Results
- High-Throughput Sequencing Analysis
- Optional Validation Data (Co-IP, Pull-down)
- Standard Timeline: 8-10 weeks
- Expedited Timeline: 6-8 weeks
Service Scope
| Bait Vector Construction We provide complete support for constructing your bait vector, including primer design, gene amplification, and validation of cloning. Our team ensures that your bait vector is fully prepared for the screening process. |
| Library Screening Our high-throughput screening process utilizes either standardized cDNA libraries or custom libraries provided by you. This approach helps us efficiently identify potential protein interactions. |
| Self-Activation Test We perform self-activation tests on bait vectors to confirm that they do not activate reporter genes independently. This step is essential for ensuring the accuracy and reliability of your screening results. |
| Positive Clone Identification We identify positive clones through PCR, sequencing, and BLAST alignment. Detailed information on interaction partners is provided to help you interpret the results. |
| Back Transformation and Validation We conduct back-transformation and validation experiments to verify the reliability of identified interactions. This ensures that the detected interactions are genuine and reproducible. |
| Customized Solutions We recognize that each research project has unique requirements. Therefore, we offer customized services tailored to your specific needs, including optimization for special protein screening and comprehensive technical support. |
Why Choose Profacgen?
High-Quality Screening with Low False Positives
Profacgen utilizes advanced yeast strains and multiple reporter genes to minimize false positives, ensuring that the interactions identified are highly reliable and relevant to your research.
Customized Solutions for Your Research Needs
We understand that every project is unique. Profacgen offers fully customized services, from bait vector construction to specialized cDNA library screening, tailored to meet your specific experimental requirements.
Expert Technical Support Throughout Your Project
Our team of experts is dedicated to supporting you from start to finish. We offer consultation on experimental design, troubleshooting, and data interpretation to ensure your project's success.
Wide Range of Ready-to-Use Libraries
We offer a diverse selection of standardized cDNA libraries for various species and tissues. If you have a custom library, we can also work with that, providing flexibility to suit your needs.
Commitment to Research Success
At Profacgen, we are dedicated to helping you achieve your research goals. Our commitment to excellence and customer satisfaction ensures that you receive the highest quality service and support.

Case Study
* NOTE: We prioritize confidentiality in our services to safeguard technology and intellectual property for enhanced future value and protection. The following case study has been shared with the client's consent.
Goal
To construct a yeast two-hybrid (Y2H) library for a specific sample provided by the customer. The library was built using RNA extracted from the provided tissues and transformed into E. coli TOP10 cells.
Results
- RNA Quality
The extracted RNA showed good quality with clear 28S and 18S bands and no degradation. The OD260/OD230 ratio was greater than 2, indicating high purity.
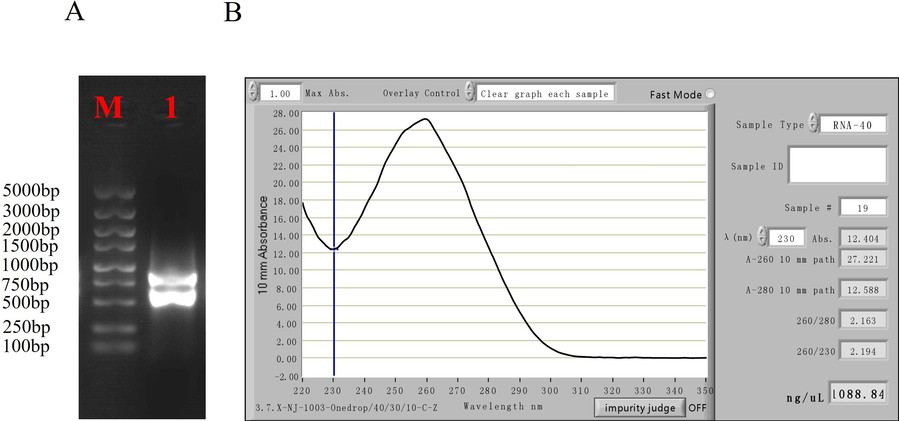 Fig2. Agarose Gel Electrophoresis and Spectrophotometer Measurement of Total RNA.
Fig2. Agarose Gel Electrophoresis and Spectrophotometer Measurement of Total RNA.
- ds cDNA Synthesis
The synthesized ds cDNA showed diffuse bands of various sizes on agarose gel electrophoresis, indicating successful synthesis.
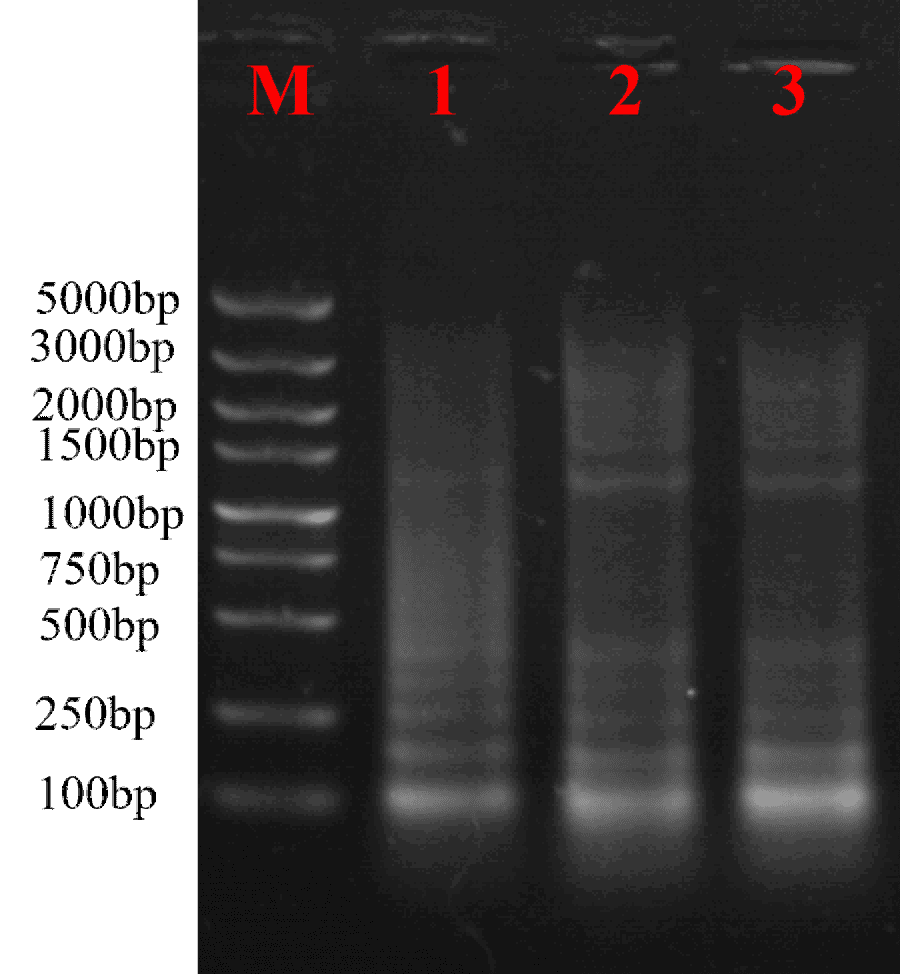 Fig3. Agarose Gel Electrophoresis of cDNA.
Fig3. Agarose Gel Electrophoresis of cDNA.
- Normalization and Small Fragment Removal
The normalized ds cDNA showed no obvious bright bands and few fragments below 500 bp, confirming successful normalization and removal of small fragments.
 Fig4. Agarose Gel Electrophoresis Detection of Homogenization and Removal of Small Fragments.
Fig4. Agarose Gel Electrophoresis Detection of Homogenization and Removal of Small Fragments.
- Clone Counting
The library capacity was determined to be xxx cfu/mL, with a total clone number of xxx.
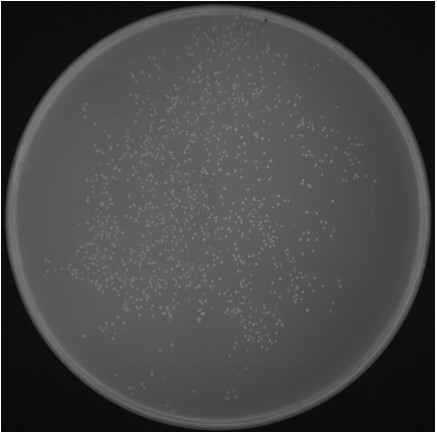 Fig5. E. coli Colony Count.
Fig5. E. coli Colony Count.
- Library Quality Identification
The average insert size of the library was approximately 1,000 bp, as confirmed by colony PCR and agarose gel electrophoresis.
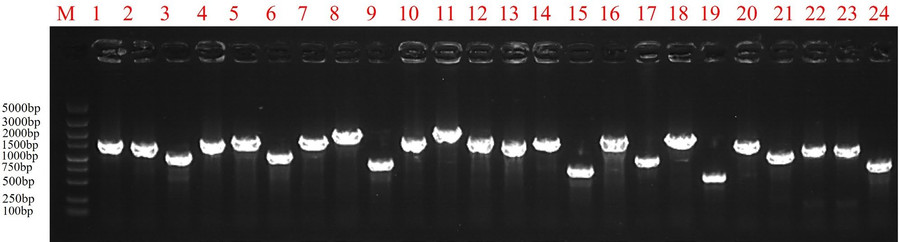 Fig6. Library quality assessment.
Fig6. Library quality assessment.
Conclusions and Discussions
The Y2H library was successfully constructed with a total CFU of xxx. The average insert size (AIS) of the library was greater than 1,000 kb. The library is suitable for downstream applications in yeast two-hybrid screening.
Goal
To verify custom protein interactions using the yeast two-hybrid (Y2H) system. Specifically, the target proteins RING-3, SNF, and MAPK were constructed into the bait vector pGBKT7, while the protein W4 was constructed into the prey vector pGADT7. The interactions between these proteins were tested by co-transforming the vectors into the AH109 yeast strain and verifying the interactions using the reporter genes HIS3 and ADE2.
Results
- Six monoclones of each transformant were randomly selected for PCR detection, and correct monoclones were preserved.
- The positive control (pGADT7-largeT & pGBKT7-p53) grew on both SD-TL and SD-TLHA plates, while the negative control (pGADT7-largeT & pGBKT7-laminC) failed to grow on the SD-TLHA plate due to its inability to activate the ADE2 and HIS3 reporter genes.
- The combinations of pGADT7-W4 with pGBKT7-RING-3, pGBKT7-SNF, and pGBKT7-MAPK could grow on both SD-TL and SD-TLHA plates, indicating interactions between W4 and RING-3, SNF, and MAPK, respectively. None of the proteins showed self-activation.
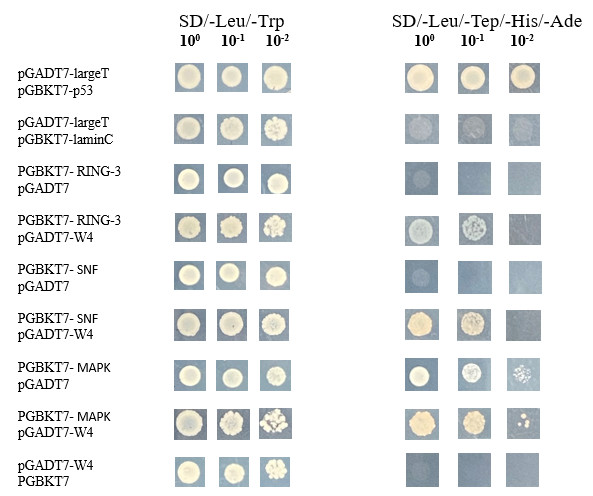 Fig8. Positive Clones Detected with HIS3 and ADE2 Reporter Genes.
Fig8. Positive Clones Detected with HIS3 and ADE2 Reporter Genes.
Conclusions and Discussions
The project was successfully completed. The interactions between W4 and RING-3, SNF, and MAPK were confirmed through the activation of the HIS3 and ADE2 reporter genes.
Customer Testimonials
"Profacgen's service was incredibly efficient. The detailed results helped us identify key protein interactions for our drug discovery project."
Dr. Emily Carter, Research Scientist at a biopharmaceutical company
"Their customized screening solutions were perfect for our unique needs. The team was responsive and the data quality exceeded expectations."
Dr. Raj Patel, Principal Investigator at a genetics research lab
"Profacgen delivered high-quality results quickly. Their expertise was crucial for our project on protein interactions."
Dr. Lisa Kim, Senior Scientist at an agricultural biotech firm
"We were impressed with the low false-positive rate and thorough validation. It made our research more efficient."
Dr. Tom Brown, Research Director at a neuroscience institute
"The detailed screening report and technical support were invaluable. Profacgen's service had a significant impact on our study."
Dr. Sarah White, Biochemist at a health research organization
"Their yeast two-hybrid screening was a game-changer for our cancer research. The precise data helped uncover new interactions."
Dr. David Lee, Research Fellow at a cancer research institute
FAQs
Resources
| Co-Immunoprecipitation (Co-IP) | Surface Plasmon Resonance (SPR) Service | Proximity-dependent Biotin Identification (BioID) Service | Yeast Display Service | Bio-Layer Interferometry (BLI) |
See more details on the principle and protocol of yeast two hybrid screening on our website. See more…

