At Profacgen, our eCLIP-Seq (enhanced Cross-Linking Immunoprecipitation Sequencing) service covers all the essentials-from prepping your samples and doing UV cross-linking and immunoprecipitation, to enriching and purifying RNA, building libraries, running the sequencing, and diving deep into data analysis. Our hands-on team will be with you every step of the way, helping you design your experiment and make sense of the data, so you get reliable, actionable results. Whether you're looking at RNA processing, hunting for miRNA targets, or figuring out how RNA-binding proteins work, our service is designed to give your research the boost it needs.
 Fig1. Schematic Diagram of EMSA.
Fig1. Schematic Diagram of EMSA.
eCLIP-Seq takes classic CLIP methods a step further, offering better resolution and efficiency for pinpointing the RNA targets of binding proteins. It mixes UV cross-linking with immunoprecipitation and high-throughput sequencing to map binding sites across the whole RNA transcriptome, covering introns, exons, and non-coding RNAs. This method really stands out from other clip-seq techniques by clearly showing where proteins interact with RNA, boosting the connection between them, and making it easier to identify binding sites across different regions like the 3'-UTR and 5'-UTR.
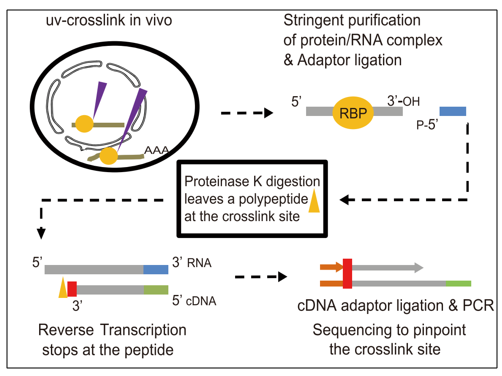 Fig2. Diagram illustrating the principle of eCLIP-seq. (Zhu.; et al. Mol Plant. 2020)
Fig2. Diagram illustrating the principle of eCLIP-seq. (Zhu.; et al. Mol Plant. 2020)
High-Resolution Mapping
eCLIP-Seq pinpoints the exact spots where RNA-binding proteins stick to the transcriptome-covering introns, exons, and non-coding RNAs-with great precision.
Enhanced Efficiency
This approach ramps up ligation efficiency while cutting down on PCR duplicate reads, which really boosts both throughput and accuracy.
Safety and Convenience
By steering clear of radioactive markers, eCLIP-Seq offers a safer, more user-friendly alternative to the traditional CLIP methods.
Detection of Transient Interactions
Thanks to UV cross-linking, it catches even fleeting RNA-protein interactions, giving you a clear picture of dynamic regulatory processes.
Comprehensive Data Analysis
Profacgen provides in-depth analysis, including quality control, sequence alignment, peak calling, GO and KEGG pathway analysis, and motif identification, so you end up with solid, usable results.
Versatile Applications
Whether you're working with common cell lines or model organisms, eCLIP-Seq adapts smoothly to a wide variety of biological samples.
Deliverables:
Service Details:
| UV Cross-Linking | Immunoprecipitation | RNA Enrichment and Purification | Library Generation and Sequencing | Data Analysis | |
| Service Content | UV exposure to form covalent bonds between RNA and proteins. | Cell lysis, RNA fragmentation, and immunoprecipitation using specific antibodies. | Purification of captured RNA fragments. | Reverse transcription, adapter ligation, qPCR, library test, and high-throughput sequencing (Illumina HiSeq or Novaseq). | Sequencing, quality control, sequence alignment, peak calling, GO and KEGG pathway analysis, and motif identification. |
| Note | Crucial for stabilizing RNA-protein complexes. | Antibody selection is critical. | Ensures high-quality RNA for sequencing. | Optimized parameters for high-quality data. | Detailed reports and visualizations provided. |
Background
The goal of this project is to deliver the eCLIP-Seq results of WT drosophila ovary and mutant X to the customer. The project involves tissue lysis, RNase digestion, immunoprecipitation, RNA library construction, sequencing, and bioinformatics analysis.
Results
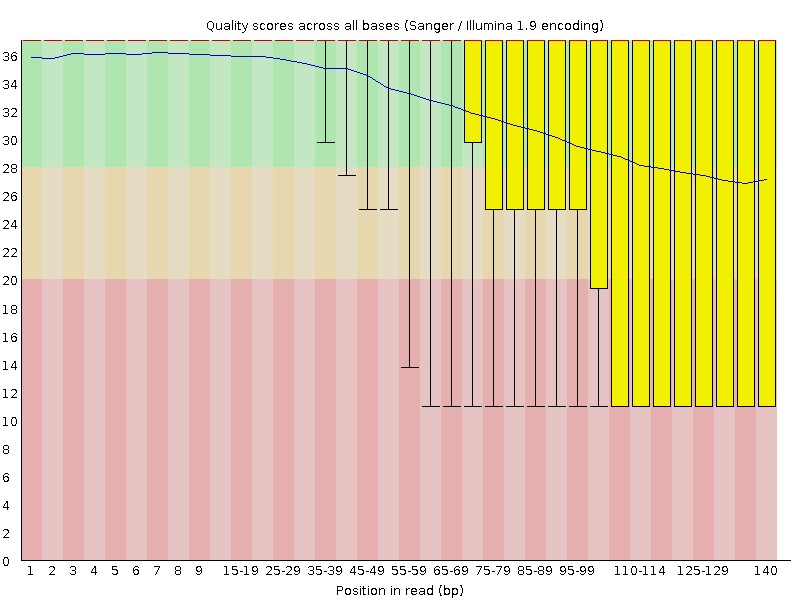 Fig3. Quality scores Per base sequence quality.
Fig3. Quality scores Per base sequence quality.
Table 1 Statistics of reference sequence alignment results.
| sample names | number of input reads | Average input read length | Mapped reads number | Mapped reads% | Average mapped length | Number of splices: Total |
| Control-Input_PE.Control-Input.NIL | 2798063 | 96 | 405572 | 14.50% | 114.92 | 10227 |
| Control-IP_PE.Control-IP.bc1 | 204205 | 127 | 80042 | 39.20% | 140.44 | 132 |
| Control-IP_PE.Control-IP.bc2 | 119849 | 134 | 34822 | 29.06% | 142.14 | 84 |
| Mutant-Input_PE.Mutant-Input.NIL | 2870779 | 125 | 990023 | 34.49% | 129.69 | 37404 |
| Mutant-IP_PE.Mutant-IP.bc1 | 344856 | 119 | 173294 | 50.25% | 129.4 | 267 |
| Mutant-IP_PE.Mutant-IP.bc2 | 160347 | 124 | 74074 | 46.20% | 131.59 | 182 |
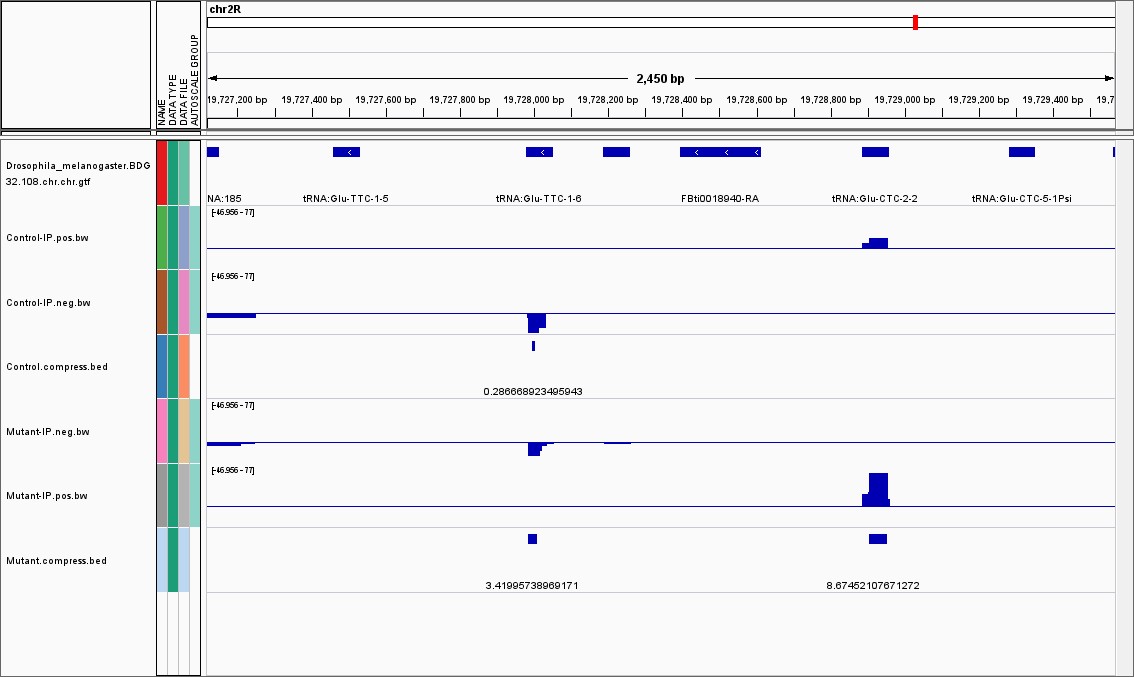 Fig4. IGV visual display.
Fig4. IGV visual display.
Table 2 Peak Calling Result Statistics.
| Sample | Peak number |
| Control.compress.bed | 125 |
| Mutant.compress.bed | 590 |
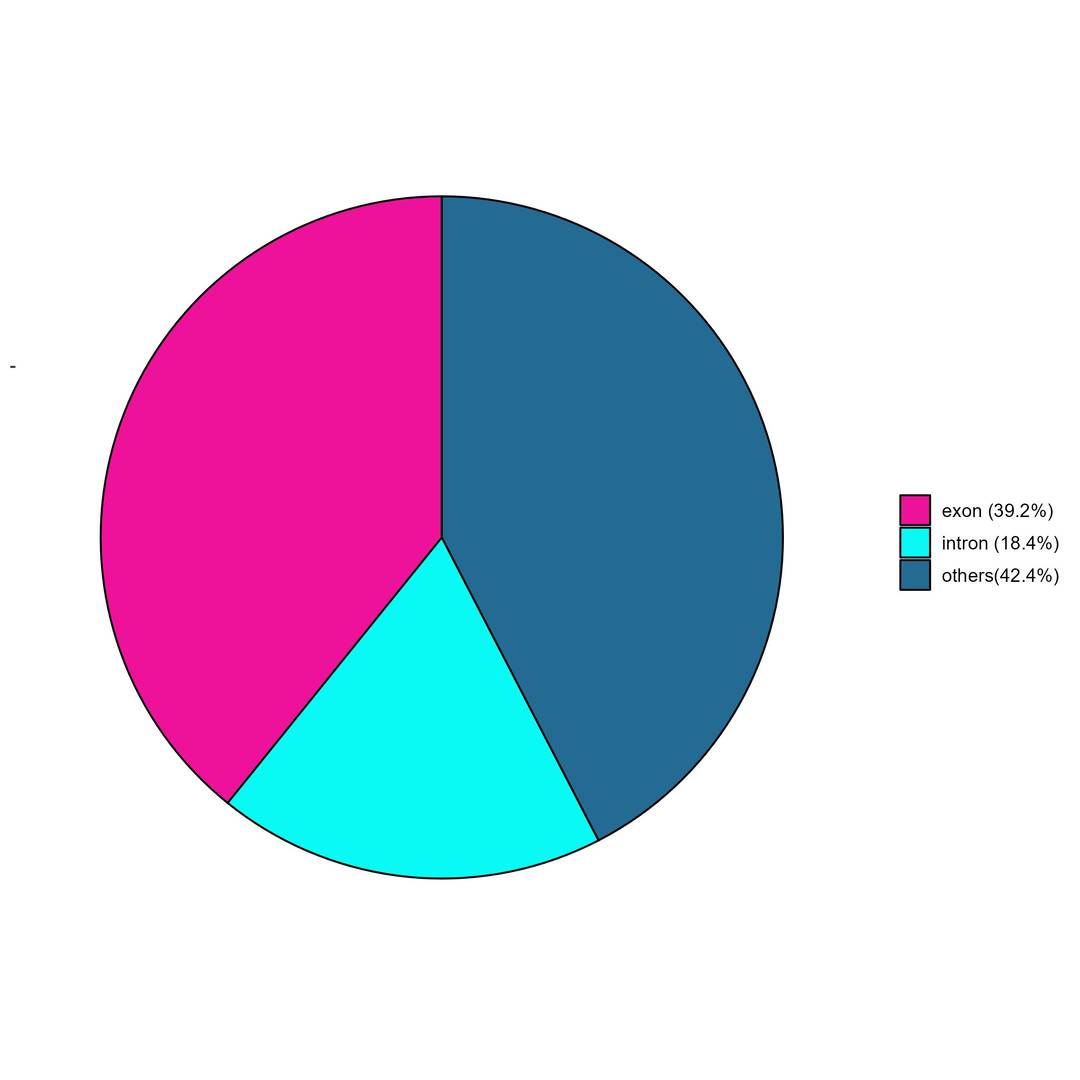 Fig5. Peak annotation distribution.
Fig5. Peak annotation distribution.
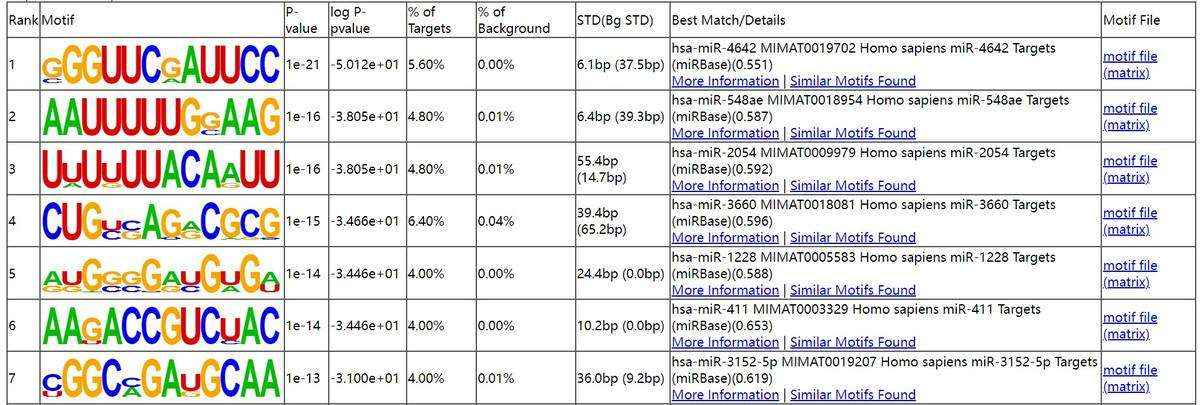 Fig6. Motif analysis.
Fig6. Motif analysis.
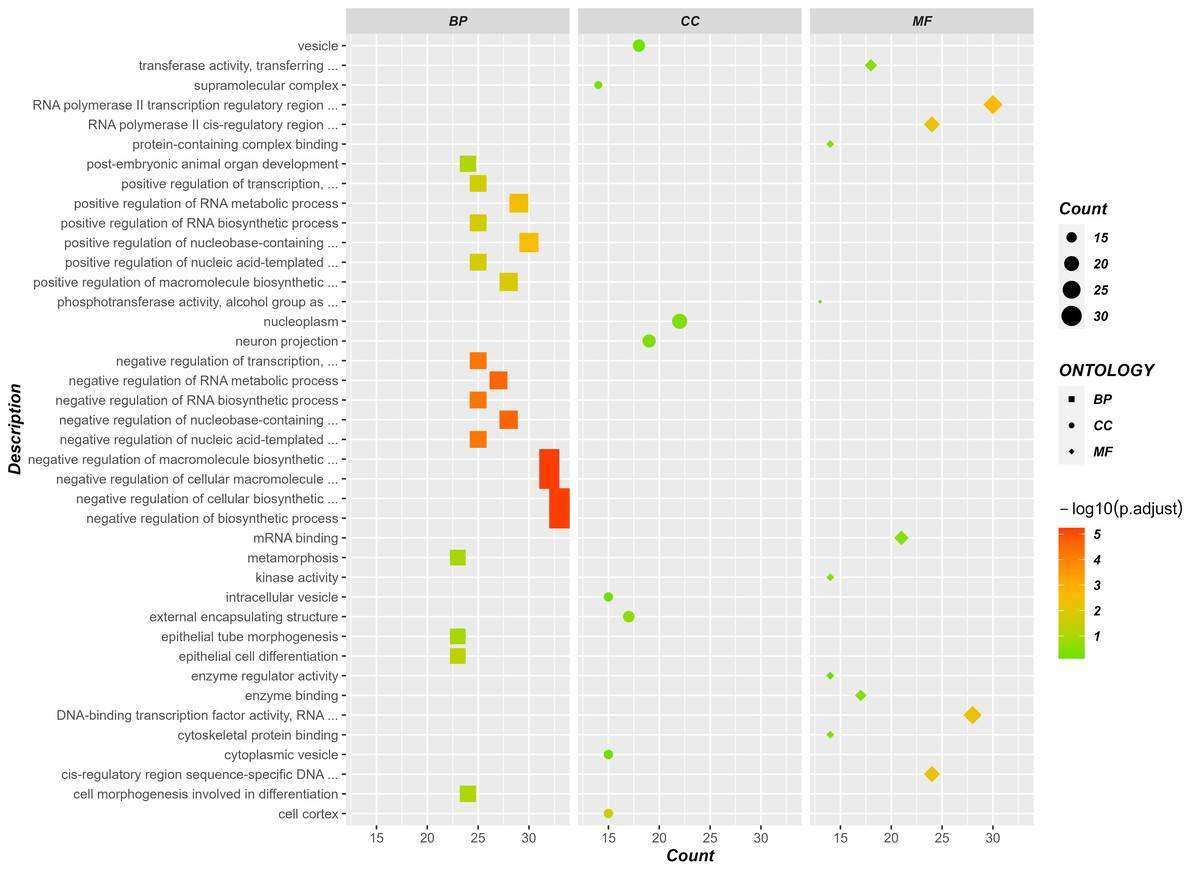 Fig7. GO enrichment analysis diagram.
Fig7. GO enrichment analysis diagram.
References:
Fill out this form and one of our experts will respond to you within one business day.