Noncoding RNA sequences, including long noncoding RNAs (lncRNAs), small nucleolar RNAs (snoRNAs), and untranslated mRNA regions, fulfill their many diverse functions through direct interactions with RNA-binding proteins (RBPs). RBPs refer to the proteins that bind to RNA, and they have powerful gene regulation capabilities. Except for a few RNAs that can function alone in the form of ribozymes, most RNAs are combined with proteins to form RNA-protein complexes. RBPs play a vital role in regulating life activities such as cell development, differentiation, metabolism, health, and disease and are essential to cellular homeostasis. Therefore, studying the interaction between RNA and protein is the key to exploring the function of RNA. Recent studies have demonstrated that hundreds of novel RBPs lack known RNA-binding domains, showing the complexity and diversity of RNA-protein complexes.
Current methods for studying RNA-protein interactions fall into two categories methods that detect proteins bound to target RNAs (RNA-centric) (shown in Fig.1) and methods that detect RNAs bound to target proteins (protein-centric). RNA-centric assays are generally divided into in vitro and in vivo. In vitro methods are particularly effective for determining which nucleotides and amino acids are involved in known RNA-protein interactions. In vivo approaches are biased towards dynamic studies such as subcellular localization, modifications, or RNA-protein interactions as protein concentrations change. Common protein-centric screens include CLIP-seq, etc., but they have the defect of poor cross-linking efficiency. Although there are many research methods for RBP, researchers are still constantly improving optimization methods to solve more biological problems.
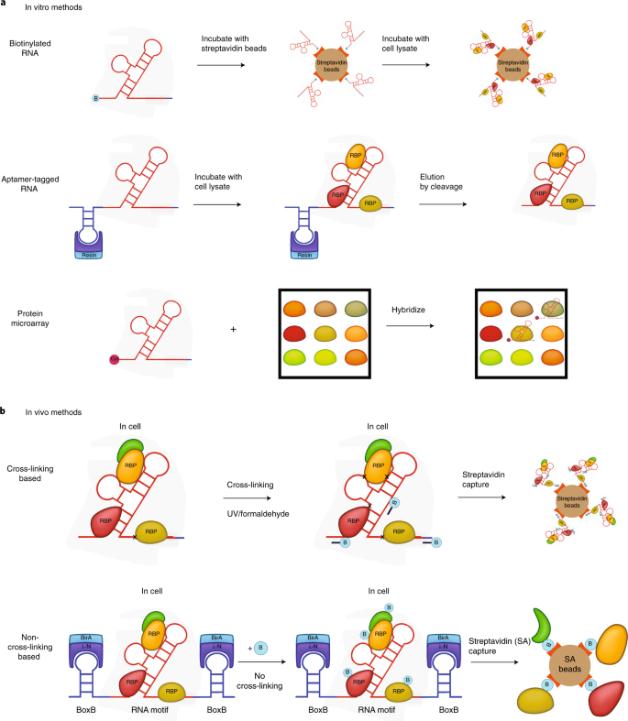 Fig.1 Schematic representation of RNA-centric methods.
Fig.1 Schematic representation of RNA-centric methods.
A new method for detecting RNA-protein binding that can be used to assess the binding interactions of RNA aptamers with their proteins on a large scale has been developed. This method can transcribe DNA directly on flow cells and measure the binding affinity of RNA aptamers to fluorescently labeled proteins. This method stops the RNA transcription of T7 RNA polymerase upstream of the Tus-bound Ter site by introducing the E. coli replication termination protein (Tus) binding sequence Ter into the system. The transcribed RNA strand remains attached to the RNA polymerase complex due to the abrupt cessation of RNA polymerase activity due to DNA-bound Tus. Then, by introducing fluorescently labeled proteins and binding to the exposed RNA aptamers, a fluorescent signal is generated that can be detected and analyzed.
Profacgen is a state-of-the-art protein service provider. We provide custom protein services in the biological sciences, enabling access to the latest tools, techniques, and expertise with competitive pricing and rapid turnaround time. We serve a broad spectrum of industrial and academic clients committed to delivering high-quality data and customer services.
Please do not hesitate to contact us for more details if you are interested in this new method, and we will provide considerate service for you. At the same time, we also offer other services and biomarker products; please move to our website for more details.
Reference
Fill out this form and one of our experts will respond to you within one business day.