DNA/RNA pull down
Background
Introduction
Getting a handle on how DNA/RNA interacts with proteins is crucial for cracking the code of gene regulation and understanding all sorts of biological activities. Basically, these interactions are key in controlling how genes get expressed. Transcription factors latch onto specific DNA bits to tweak gene activity, and RNA-binding proteins step in for things like RNA stability and splicing. However, older methods like ChIP-seq and RIP can be a hassle-they need specific antibodies, which aren't always easy to find, especially for rare proteins or non-standard species, and involve complex steps that might not deliver top-notch data. That's where DNA/RNA pull-down comes in-a game-changer that ditches the antibodies and uses biotin-labeled probes with streptavidin beads to nab DNA/RNA-protein combos. This makes it a high-throughput, super flexible method that's great for exploring and validating interactions across various species and setups.
What is DNA/RNA pull down?
DNA/RNA pull-down is a great go-to method for grabbing proteins that connect with certain DNA or RNA sequences. Here's the deal: you start by creating DNA or RNA probes with biotin, which pair with the sequences you're interested in. These probes get attached to magnetic beads coated with streptavidin, which really loves biotin. You then mix this DNA/RNA-bead combo with a cell lysate, letting proteins that specifically stick to your DNA/RNA bind to the beads. After giving everything a good wash to clear away any proteins that don't belong, you elute the DNA/RNA-protein pairs from the beads. You can then break these down further using techniques like Western blot to spot known proteins or mass spectrometry to figure out any unknown ones.
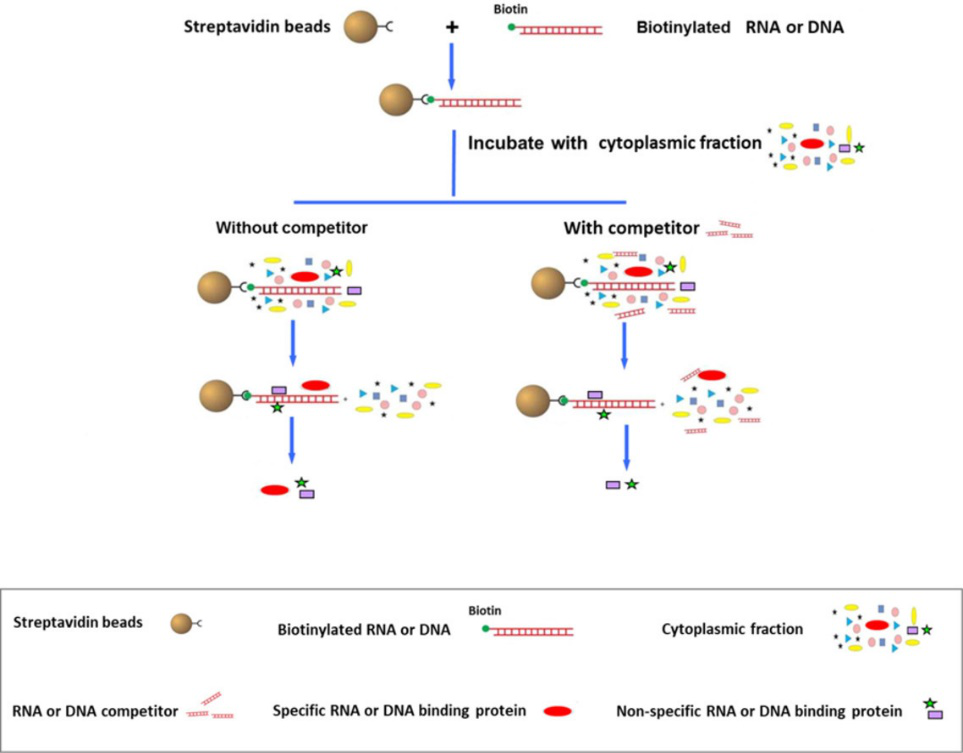 Fig1. DNA/RNA pull down overview.
Fig1. DNA/RNA pull down overview.
Applications of DNA/RNA pull down
Uses for DNA/RNA pull-down stretch across molecular biology and biotech fields. It's especially handy for:
- Screening or Validating Binding Proteins: Pinpointing and confirming proteins that link up with particular DNA or RNA sequences.
- Finding Transcription Factors: Spotting new transcription factors that attach to specific gene promoters, shedding light on how genes are regulated.
- Protein-Protein Interaction Networks: Checking out how RNA-binding proteins interact with other cell parts to better understand post-transcriptional regulatory pathways.
- Drug Screening: Looking for possible drug targets by finding proteins that connect with certain nucleic acid sequences, paving the way for new treatments.
Service Procedure
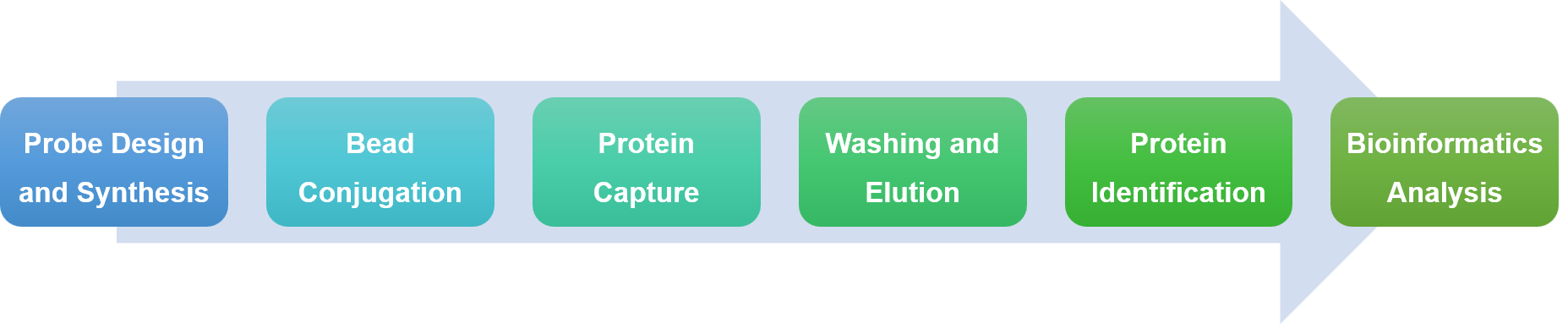
At Profacgen, we provide full DNA/RNA pull-down services to help you pinpoint and confirm DNA/RNA-binding proteins with great accuracy. Our package covers everything from probe design and protein capture to mass spectrometry analysis and thorough bioinformatics support.
| Probe Design and Synthesis | Protein Capture | Protein Identification | Bioinformatics Analysis | |
| Description | Make biotin-labeled DNA/RNA probes that match your target sequences. These probes are key for grabbing proteins connected to the nucleic acids you're interested in. | Mix the DNA/RNA-bead combo with cell lysates to catch interacting proteins. This lets proteins that specifically latch onto your target sequences stick to the beads. | Use mass spectrometry to find the proteins you've captured and confirm interactions with a Western blot. Mass spectrometry gives you detailed info about the proteins binding to your targets, and Western blot helps confirm specific ones are there. | Provide detailed analysis of identified proteins, including functional annotation and pathway analysis. This step helps in understanding the biological significance of the identified interactions. |
| Key Points |
|
|
|
|
Why Choose Profacgen?
- Expertise and Experience: Profacgen's team has extensive experience in nucleic acid-protein interaction studies, ensuring high-quality results.
- Comprehensive Support: We offer full guidance on experimental design, including upstream and downstream validation methods.
- In-House Proteomics Platform: Our advanced proteomics platform ensures accurate mass spectrometry identification and in-depth bioinformatics analysis.
- Ongoing Support: Our technical support continues until project completion, ensuring smooth downstream experiments and publication support.
Case Study
Background
The service is for screening of interactive proteins of target DNA in one kind of primary cells, by DNA pull down assay. Briefly, the target DNA gene was synthesized and labeled with biotin and immobilized onto streptavidin agarose bead. After incubation with total protein extracts, the bound proteins were eluted and then detected by mass spectrometry.
Results
- Gene Synthesis: The DNA sequence of the target gene was synthesized and validated by sequencing.
- Agarose Gel Electrophoresis: The biotin-labeled and unlabeled probes were tested, confirming the correct size of the DNA fragments.
- SDS-PAGE for Proteins: The SDS-PAGE results showed distinct protein bands in the pull-down product obtained with biotin-labeled DNA probes, indicating successful protein capture.
- Mass Spectrometry: Mass spectrometry identified specific binding proteins in the experimental group.
- Bioinformatic Analysis:
- GO Annotation: Differential proteins were classified according to the three ontologies using QuickGO.
- KEGG Pathway Analysis: Identified pathways involved in the biological functions of the proteins.
- String Analysis: Interactions of differential proteins were analyzed, with detailed interaction information provided.
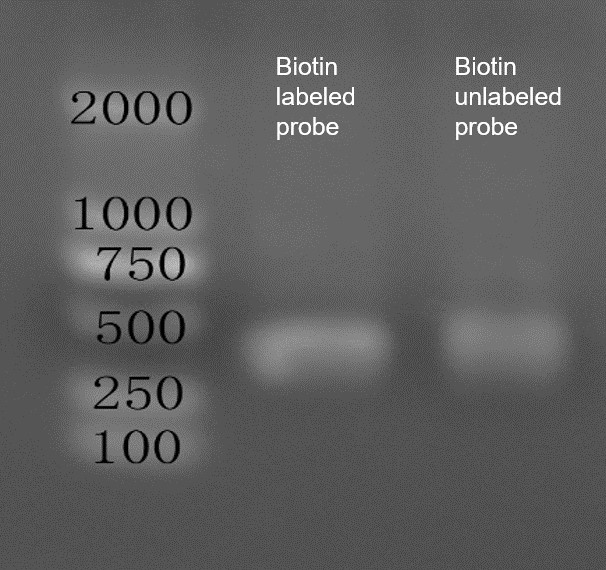 Fig2. Agarose Gel Electrophoresis of DNA Probes.
Fig2. Agarose Gel Electrophoresis of DNA Probes.
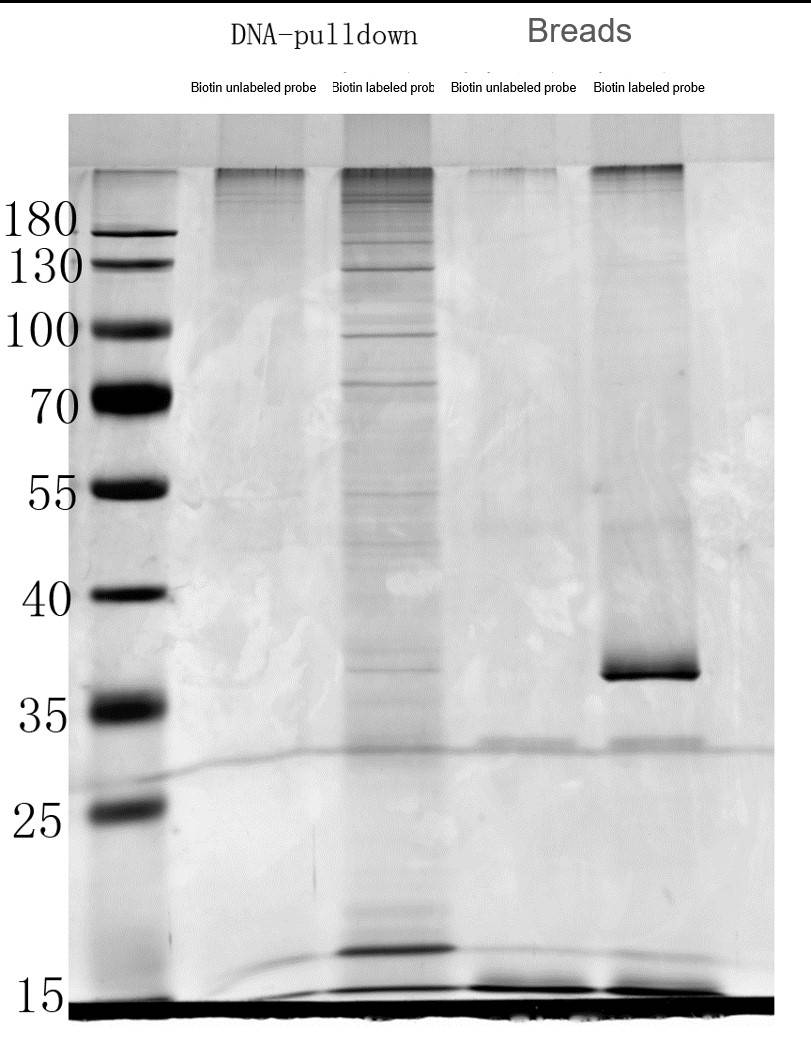 Fig3. The SDS-PAGE Results for Proteins by DNA Pull Down.
Fig3. The SDS-PAGE Results for Proteins by DNA Pull Down.
Background
The service quotation is for screening of interactive proteins of target lncRNA in human pulmonary artery smooth muscle cells (primary cells), by RNA pull down assay. Briefly, the target lncRNA gene was synthesized and transcribed in vitro, labeled with biotin and immobilized onto streptavidin agarose bead. After incubation with total protein extracts, the bound proteins were eluted and then detected by mass spectrometry.
Results
- Gene Synthesis: The DNA sequence of the target lncRNA gene was synthesized and validated by sequencing.
- Obtaining of Probes through in vitro Transcription: Sense and anti-sense biotin-labeled probes were obtained by in vitro transcription, with correct DNA fragment sizes confirmed.
- Silver Staining for Proteins: Silver staining results showed distinct differences between the Sense and Anti-sense pull-down results, with multiple protein bands observed in the Sense pull-down.
- Mass Spectrometry: Mass spectrometry identified 119 specific binding partners in the experimental group.
- Bioinformatic Analysis:
- GO Annotation: Differential proteins were classified according to the three ontologies using QuickGO.
- KEGG Pathway Analysis: Identified pathways involved in the biological functions of the proteins.
- String Analysis: Interactions of differential proteins were analyzed, with detailed interaction information provided.
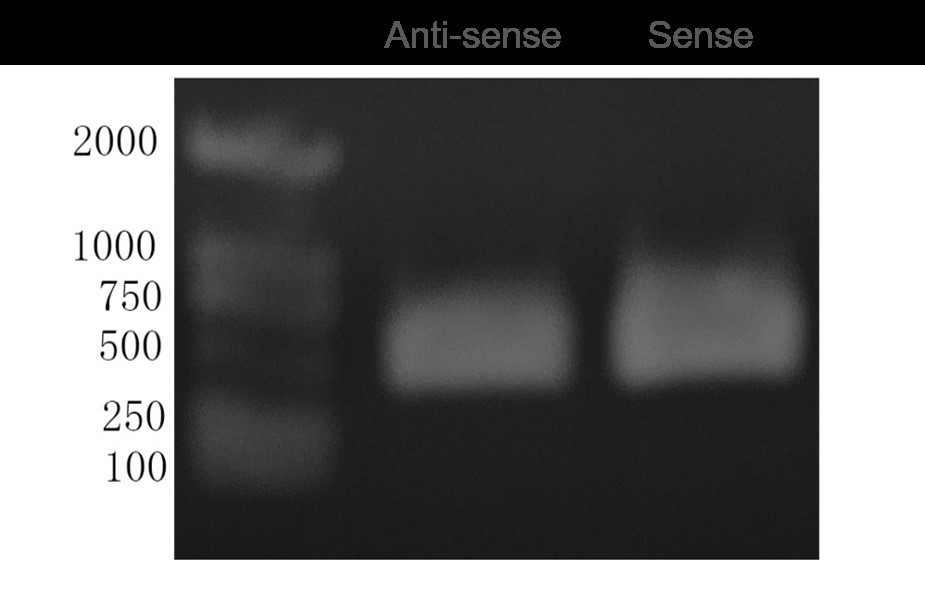 Fig4. Agarose Gel Electrophoresis of Sense and Anti-sense Amplified DNA Fragments.
Fig4. Agarose Gel Electrophoresis of Sense and Anti-sense Amplified DNA Fragments.
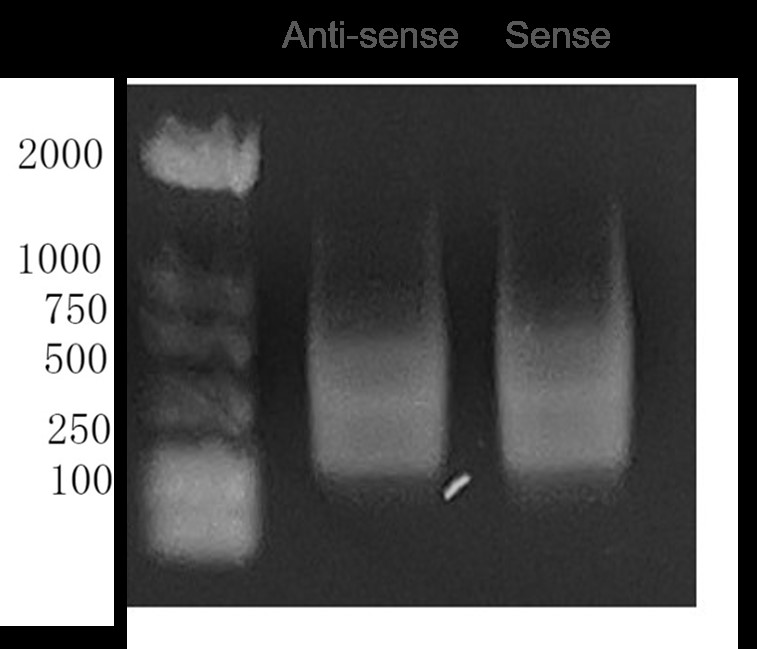 Fig5. Agarose Gel Electrophoresis of Sense and Anti-sense DNA Fragments by in vitro Transcription.
Fig5. Agarose Gel Electrophoresis of Sense and Anti-sense DNA Fragments by in vitro Transcription.
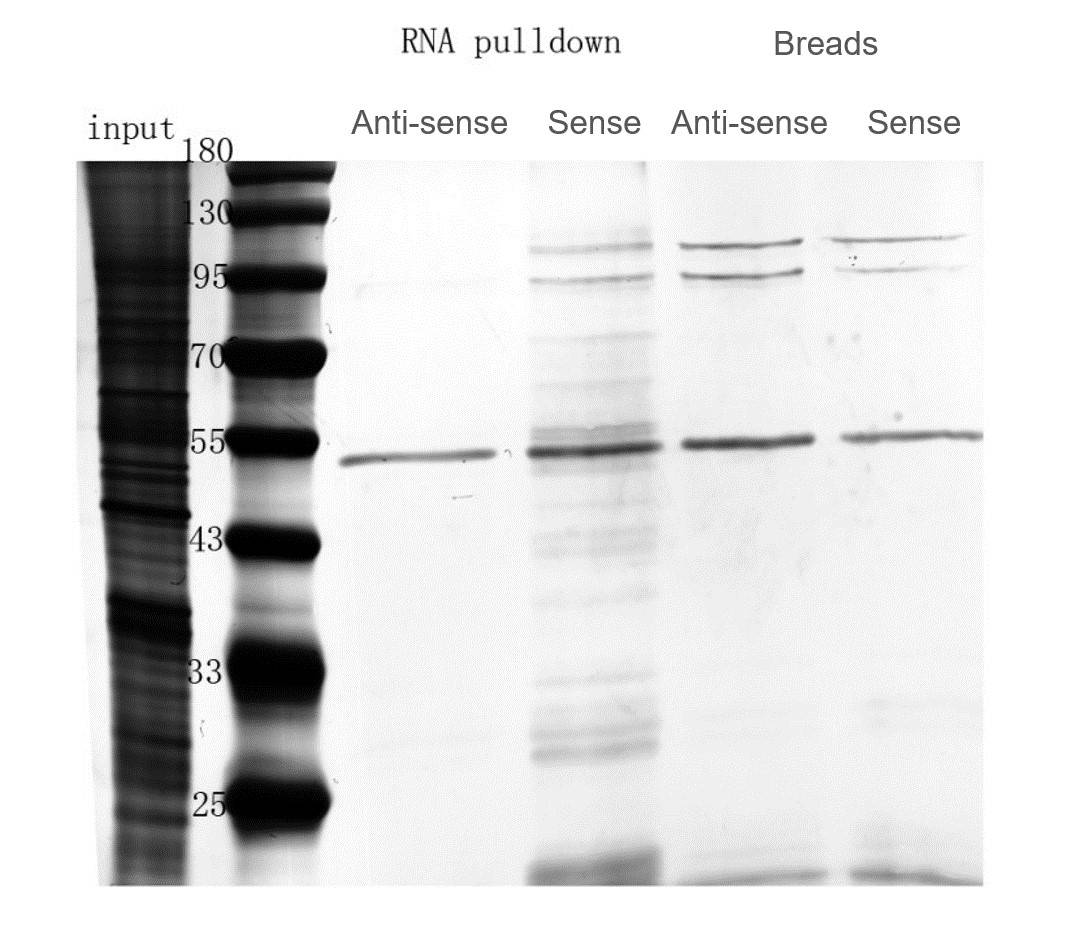 Fig6. The Silver Staining Results for Proteins on SDS-PAGE.
Fig6. The Silver Staining Results for Proteins on SDS-PAGE.
FAQs
Resources
References:
- Sui H.; et al. A pull-down assay using DNA/RNA-conjugated beads with a customized competition strategy: An effective approach to identify DNA/RNA binding proteins. MethodsX. 2020;7:100890.
- Tsuji Y.; et al. Optimization of Biotinylated RNA or DNA Pull-Down Assays for Detection of Binding Proteins: Examples of IRP1, IRP2, HuR, AUF1, and Nrf2. Int J Mol Sci. 2023;24(4):3604.
- Sui H, Imamichi T. A DNA Pull-Down Assay with Diversity Forms of Competitor for Detecting or Evaluating Protein-DNA Interactions. Methods Mol Biol. 2023;2599:1-10.