MeRIP-Seq, also known as Methylated RNA Immunoprecipitation Sequencing, is a modern technique that blends RNA-protein co-immunoprecipitation with high-throughput sequencing to provide a detailed view of RNA methylation throughout the transcriptome. This approach helps researchers untangle the dynamic patterns of RNA methylation, offering fresh perspectives on processes like cell differentiation, development, and disease progression. At Profacgen, we deliver top-notch MeRIP-Seq services aimed at clearing up the complexities of RNA methylation and propelling your research forward.
MeRIP-Seq is an effective technique for detecting and measuring RNA methylation across the entire transcriptome. First, RNA is extracted from samples and chopped into smaller pieces to make it more manageable. Then, specialized antibodies are used to single out the methylated parts, capturing modifications like m6A, m5C, and others. The isolated RNA fragments are then turned into libraries for sequencing with high-throughput systems. Finally, advanced bioinformatics tools help map where the methylation occurs, measure its level, and detect any changes in methylation patterns.
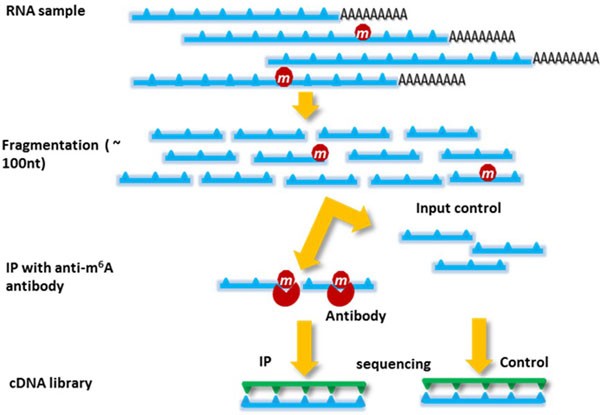 Fig1. Diagram illustrating the principle of MeRIP-seq. (Cui.; et al. BMC Genomics. 2015)
Fig1. Diagram illustrating the principle of MeRIP-seq. (Cui.; et al. BMC Genomics. 2015)
MeRIP-Seq stands out with its precision and effectiveness. It uses specific antibodies to latch onto methylated RNA, ensuring the methylation sites are detected correctly without many errors. It covers the whole transcriptome, which means it looks at all RNA at once, allowing researchers to find new methylation spots and see the big picture of RNA changes.
When it comes to measuring methylation levels, MeRIP-Seq is spot on, making it easier to compare different conditions and find areas with varying methylation intensities. By connecting methylation patterns to gene actions and what's happening in the cell, it sheds light on how RNA methylation controls biological processes like growth, response to stress, and disease evolution.
Lastly, MeRIP-Seq easily works alongside other data collection methods like RNA-Seq and proteomics. This teamwork offers a deeper dive into how genes are regulated and how biological systems function.
Cancer Research
MeRIP-Seq plays a vital role in studying the impact of RNA methylation on cancer. It uncovers the genes and pathways that m6A methylation affects, which are linked to how tumors grow, spread, and resist treatment.
Development and Differentiation
This technique is used to monitor changes in RNA methylation during early stages of embryo development, tissue growth, and the transformation of stem cells into different cell types. It provides insights into how m6A influences gene activation and cell destiny.
Stress Response
MeRIP-Seq finds changes in m6A across the transcriptome under stress like heat shock or DNA damage. It shows how RNA methylation adjusts under stress, helping us understand gene regulation during tough times.
Gene Expression Regulation
By mapping where m6A appears on RNA, MeRIP-Seq uncovers how methylation impacts mRNA stability, breakdown, splicing, and translation. It helps pinpoint functional features tied to m6A modifications.
Circular RNA Translation
This approach looks into the methylation of circular RNAs and their part in managing translation, helping us learn more about what these unique RNAs do inside cells.
RNA Metabolism
MeRIP-Seq takes a closer look at how methylation impacts RNA metabolism-like mRNA stability, its breakdown, and where it settles in the cell-to better understand RNA's role and behavior in cell functions.

MeRIP-Seq sample requirements:
meRIP-Deq analysis project:
| Fundamental Analysis | Advanced Analysis |
| Remove adapters and quality control QC. | Association analysis (with RNA-seq or multi-omics association analysis) |
| Reference Genome Alignment (Mapping) | Association Analysis Heatmap (Heatmap) |
| Enriched region identification (Peakcalling) | IGV peak diagram |
| Annotation of enriched regions (PeakingAnno) | |
| Motif Analysis | |
| Peak gene GO and KEGG analysis. | |
| Differential Peak analysis (differential RNA modification mRNA, lncRNA) | |
| Differential Peak gene GO and KEGG analysis. |
Optional services: Single-Cell and Spatial Transcriptome MeRIP-Seq
Plus, we're offering single-cell and spatial transcriptome MeRIP-Seq services. With single-cell MeRIP-Seq, you can dive into RNA methylation modifications in individual cells, which is great for exploring cell differences. On the other hand, spatial transcriptome MeRIP-Seq maps out where RNA methylation is happening in tissues, aiding in understanding tissue development and disease mechanisms.
Background
The primary objective of this project was to identify and characterize m6A modifications in target cells using MeRIP-seq. The aim was to understand the distribution and functional impact of m6A modifications by analyzing RNA fragments enriched with m6A antibodies and comparing them to input controls. This project aimed to provide insights into the regulatory roles of m6A in gene expression and cellular function.
Results
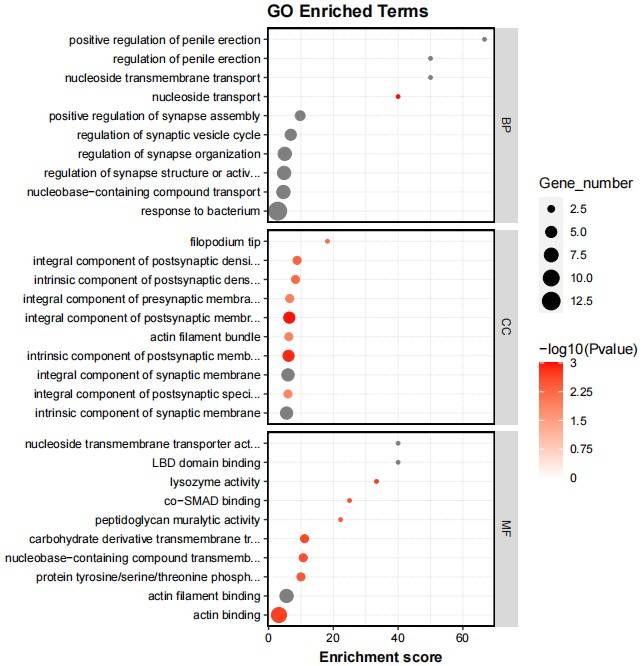 Fig2. GO Enrichment Analysis of Differential Genes for Control Group.
Fig2. GO Enrichment Analysis of Differential Genes for Control Group.
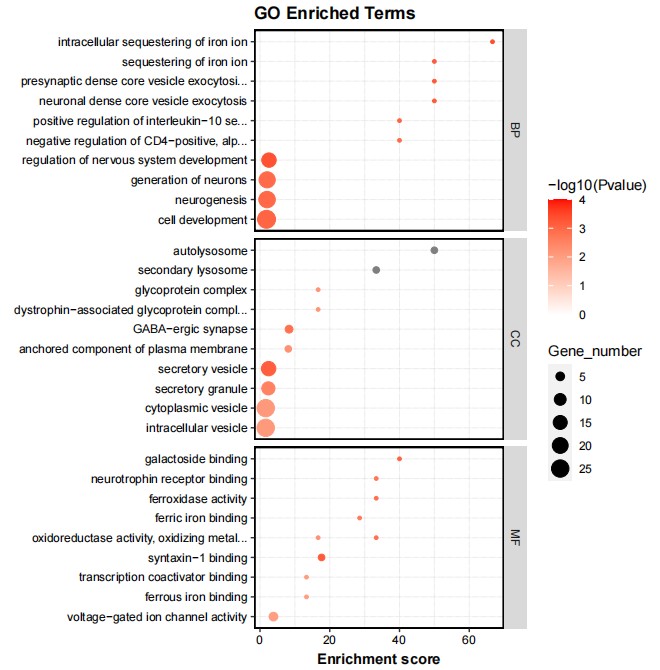 Fig3. GO Enrichment Analysis of Differential Genes for Knockout Group.
Fig3. GO Enrichment Analysis of Differential Genes for Knockout Group.
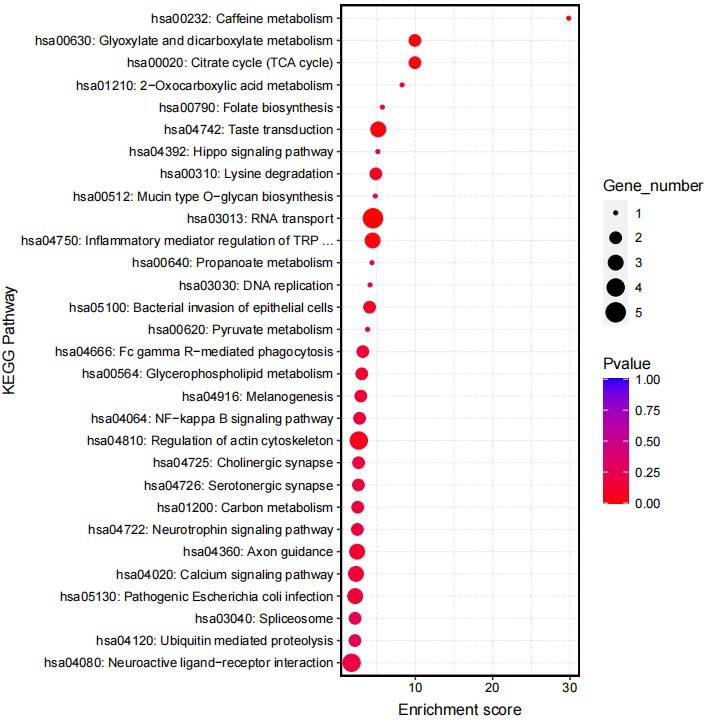 Fig4. KEGG Analysis of Differential Genes for Control Group.
Fig4. KEGG Analysis of Differential Genes for Control Group.
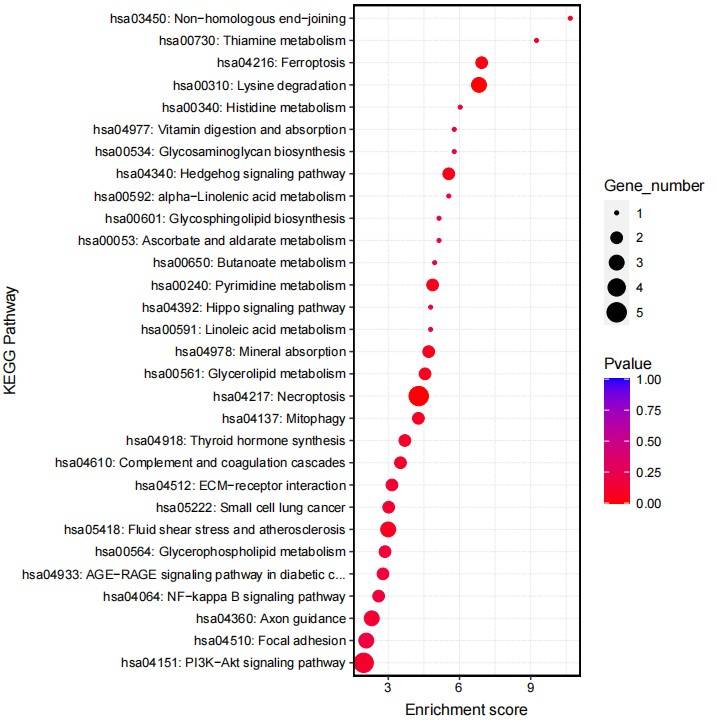 Fig5. KEGG Analysis of Differential Genes for Knockout Group.
Fig5. KEGG Analysis of Differential Genes for Knockout Group.
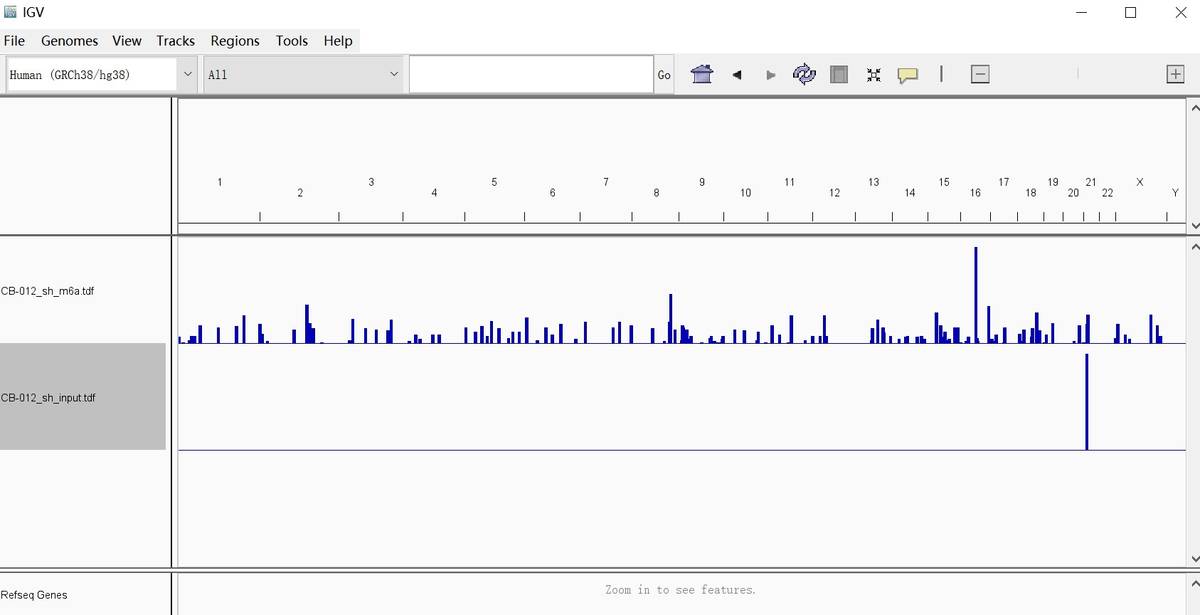 Fig6. Visualization Results.
Fig6. Visualization Results.
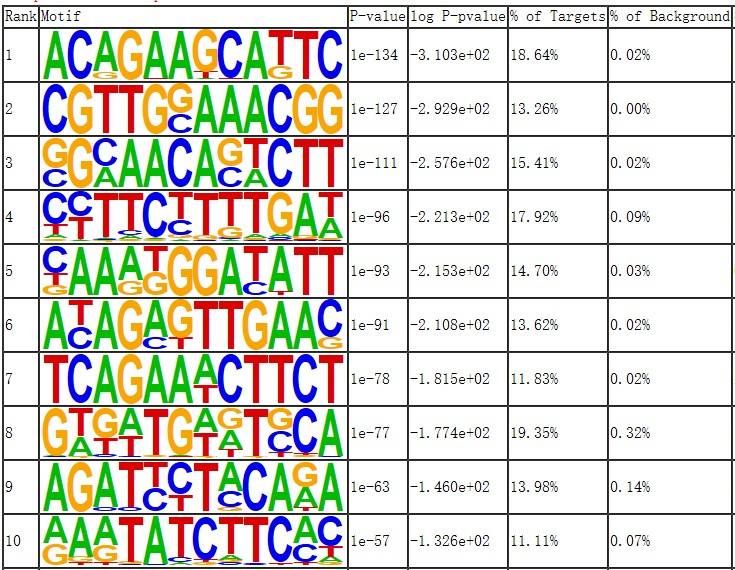 Fig7. Motif Analysis of Differential Genes for Control Group.
Fig7. Motif Analysis of Differential Genes for Control Group.
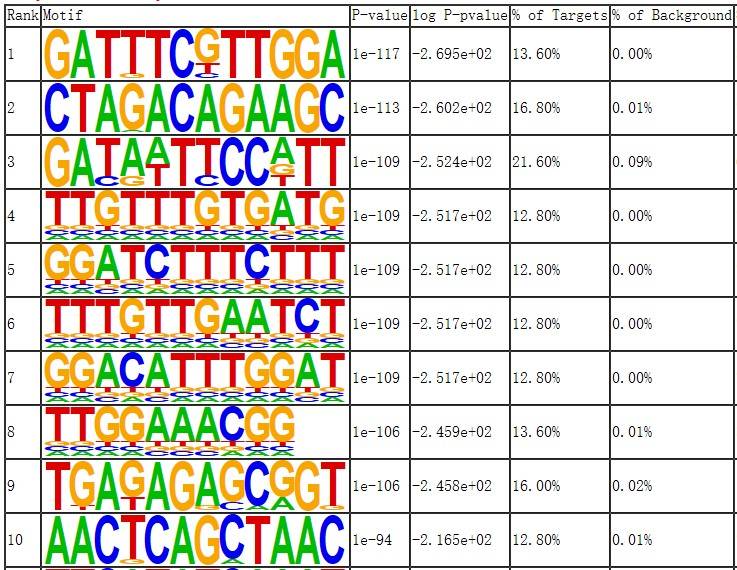 Fig8. Motif Analysis of Differential Genes for Knockout Group.
Fig8. Motif Analysis of Differential Genes for Knockout Group.
References:
Fill out this form and one of our experts will respond to you within one business day.