CLIP-Seq Service
Background
Introduction
Profacgen's got a solid CLIP-Seq (Cross-Linking Immunoprecipitation Sequencing) service to help researchers dig into RNA-protein interactions and how genes get regulated after they're transcribed. This service uses the latest techniques to map out where RNA-binding proteins (RBPs) connect across the whole genome, giving you a glimpse into the networks that manage gene expression. We handle everything: from getting your samples ready and doing the UV cross-linking to immunoprecipitation, high-throughput sequencing, and breaking down the data. This way, you get reliable results and insights that really matter for your research.
What is CLIP-Seq?
CLIP-Seq, or HITS-CLIP, is a go-to method for figuring out RNA-protein interactions and where RNA latches onto proteins. The process starts by zapping RNA-protein complexes with UV light to freeze them in place. Then, they pull out these complexes using an antibody that's tuned to the protein they're interested in. After that, they sequence the cross-linked RNA bits to pin down exactly where the proteins are hanging out. Profacgen's irCLIP-Seq technology enhances this process by using infrared fluorescent dye instead of traditional radioactive materials, improving safety and broadening application areas. Additionally, our use of thermostable group II intron reverse transcriptase for cDNA synthesis improves experimental efficiency and simplifies the technical procedure.
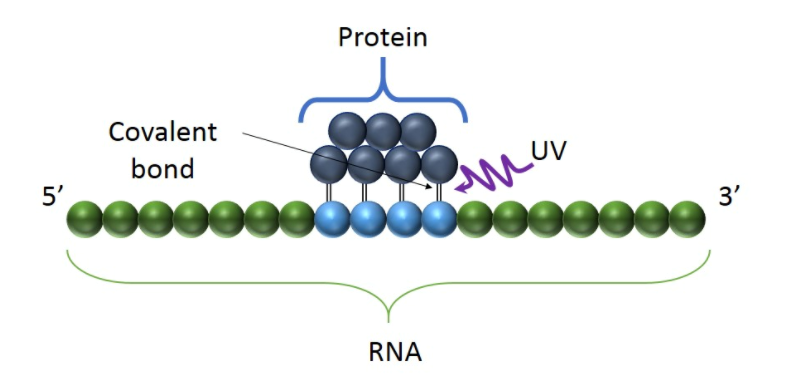 Fig1. UV light creates covalent bonds at direct RNA-protein contact points, preserving their interactions.
Fig1. UV light creates covalent bonds at direct RNA-protein contact points, preserving their interactions.
Applications of CLIP-Seq
CLIP-Seq is widely used in molecular biology and genetics to explore gene regulation after transcription:
RNA-Protein Interaction: It helps find where RNA-binding proteins grab onto RNA, showing how they control gene activity.
IncRNA/circRNA Roles: You can check how long non-coding RNA and circular RNA interact with proteins, revealing their part in gene regulation and therapy possibilities.
miRNA Targets: CLIP-Seq tracks where Argonaute proteins attach, helping us learn how miRNAs control gene activity.
Mapping RNA-Protein Networks: It provides a wide view of where RNA and proteins bind across all RNA transcripts, key for grasping how cells react to signals.
Disease Insights: By pinpointing RNA-protein interactions tied to diseases, CLIP-Seq aids in crafting new treatments and identifying possible drug targets.
Service Procedure
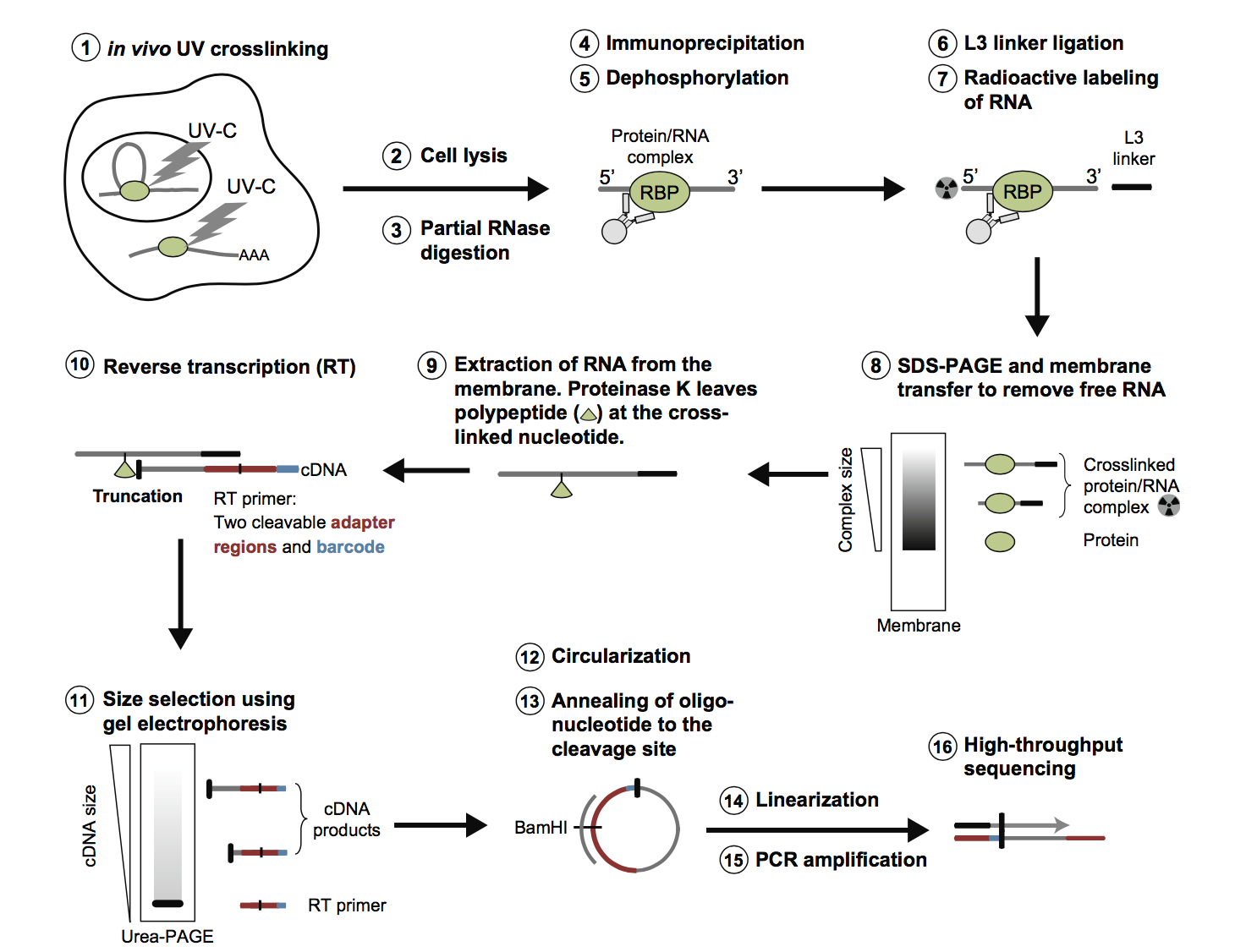 Fig2. Workflow of CLIP-Seq at Profacgen. (Huppertz.; et al. Methods. 2014)
Fig2. Workflow of CLIP-Seq at Profacgen. (Huppertz.; et al. Methods. 2014)
Sample Submission and Data Readout
UV cross-linked cell sample is recommended for its stability (107 cells is preferred). Live cells in culture medium are also acceptable.
Sequencing analysis will be Single End 50 cycle, 20-40M.
A comprehensive set of data analysis includes quality control of reads, sequence alignment, DNA element analysis, Reads allocation in transcriptome, Reads allocation around TSS/TTS/Start Codon/Stop Codon, PEAK annotation, GO (gene ontology) annotation and classification, KEGG pathway analysis, etc.
Why Choose Profacgen?
- State-of-the-Art Technology: Utilize the latest irCLIP-Seq technology for high sensitivity and safety.
- Comprehensive Analysis: Provide detailed data analysis and reporting to support your research.
- Expert Support: Our experienced team will design and optimize your CLIP project, ensuring high flexibility and efficiency.
- Customized Solutions: Tailored services to meet your specific research needs.
Case Study
Background
The aim of this project is to screen for interactions between rRNA and other RNAs, such as mRNA of X protein in target cells, using CLIP-Seq. The RNA-X complex was separated using X antibody, and the bound RNAs were analyzed by high-throughput sequencing to identify binding sites and understand the RNA-protein interaction landscape.
Results
Sequence Alignment and Coverage Statistics:
- Clean reads were aligned to the reference genome with high coverage and depth.
- Unique reads were used for downstream analysis.
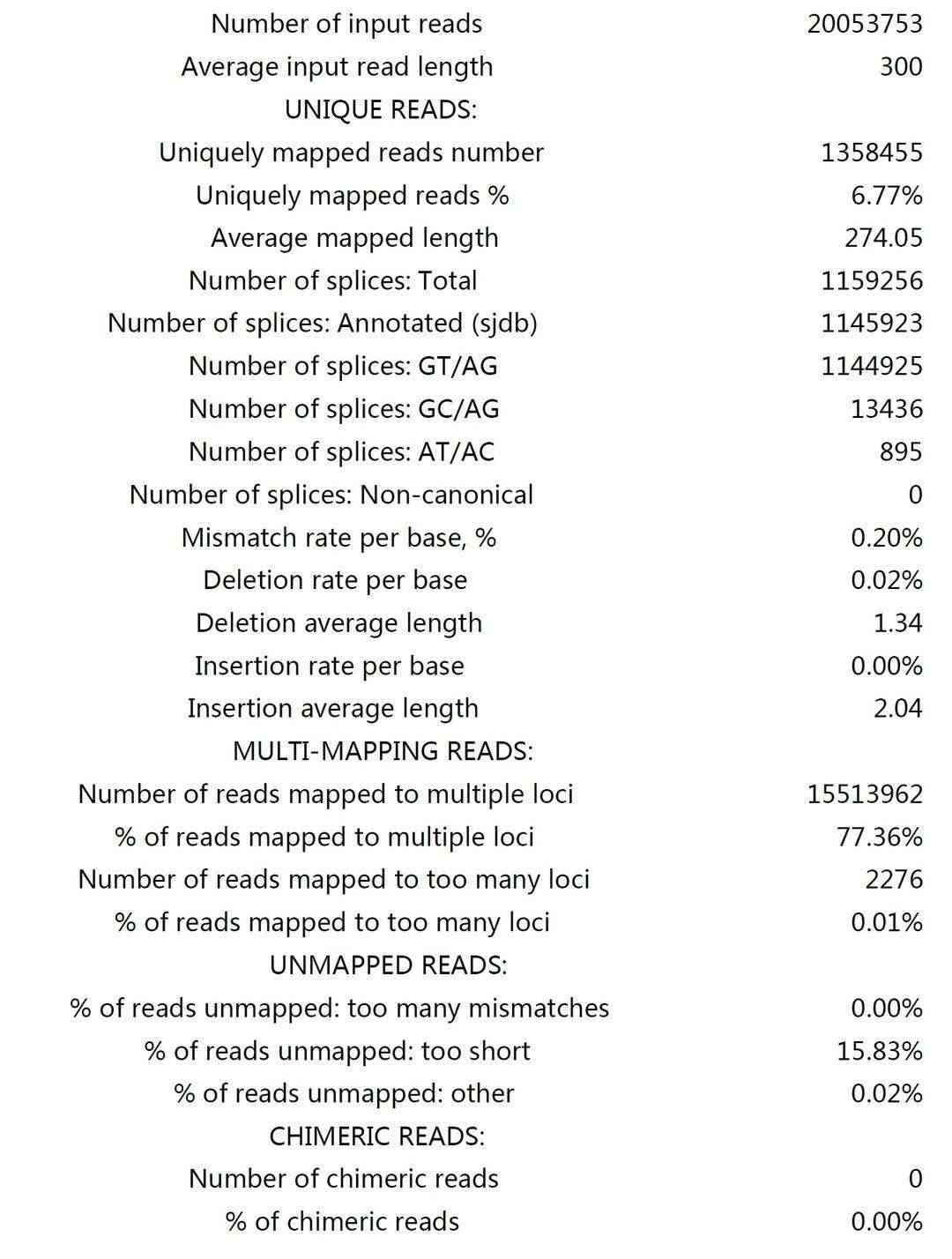 Fig3. X_CLIP Sample.
Fig3. X_CLIP Sample.
Binding Peak Identification:
- A total of 3,302 binding sites were identified for X protein.
- Binding peaks were annotated and visualized using IGV tools.
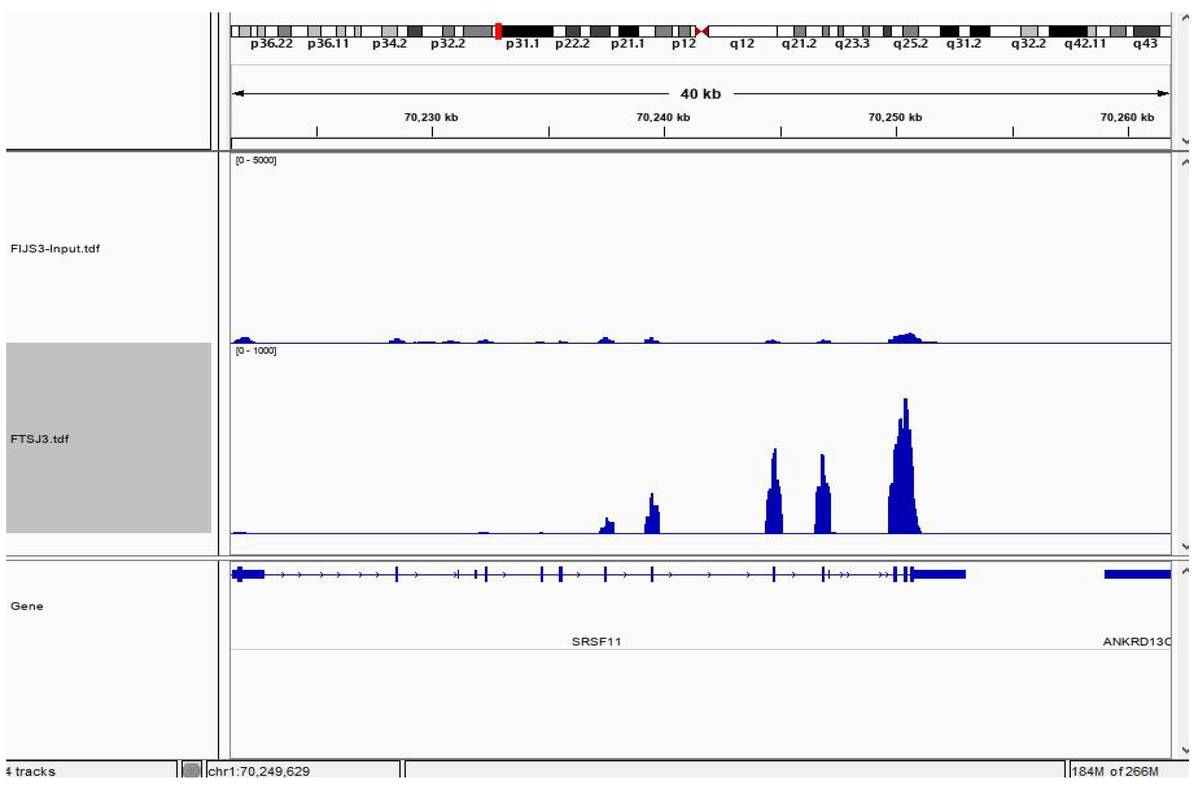 Fig4. CLIP-seq Result Visualization.
Fig4. CLIP-seq Result Visualization.
KEGG Pathway Analysis:
- Differential peak-related genes were analyzed for KEGG pathway enrichment.
- Significant pathways were identified, providing insights into the biological functions regulated by X protein.
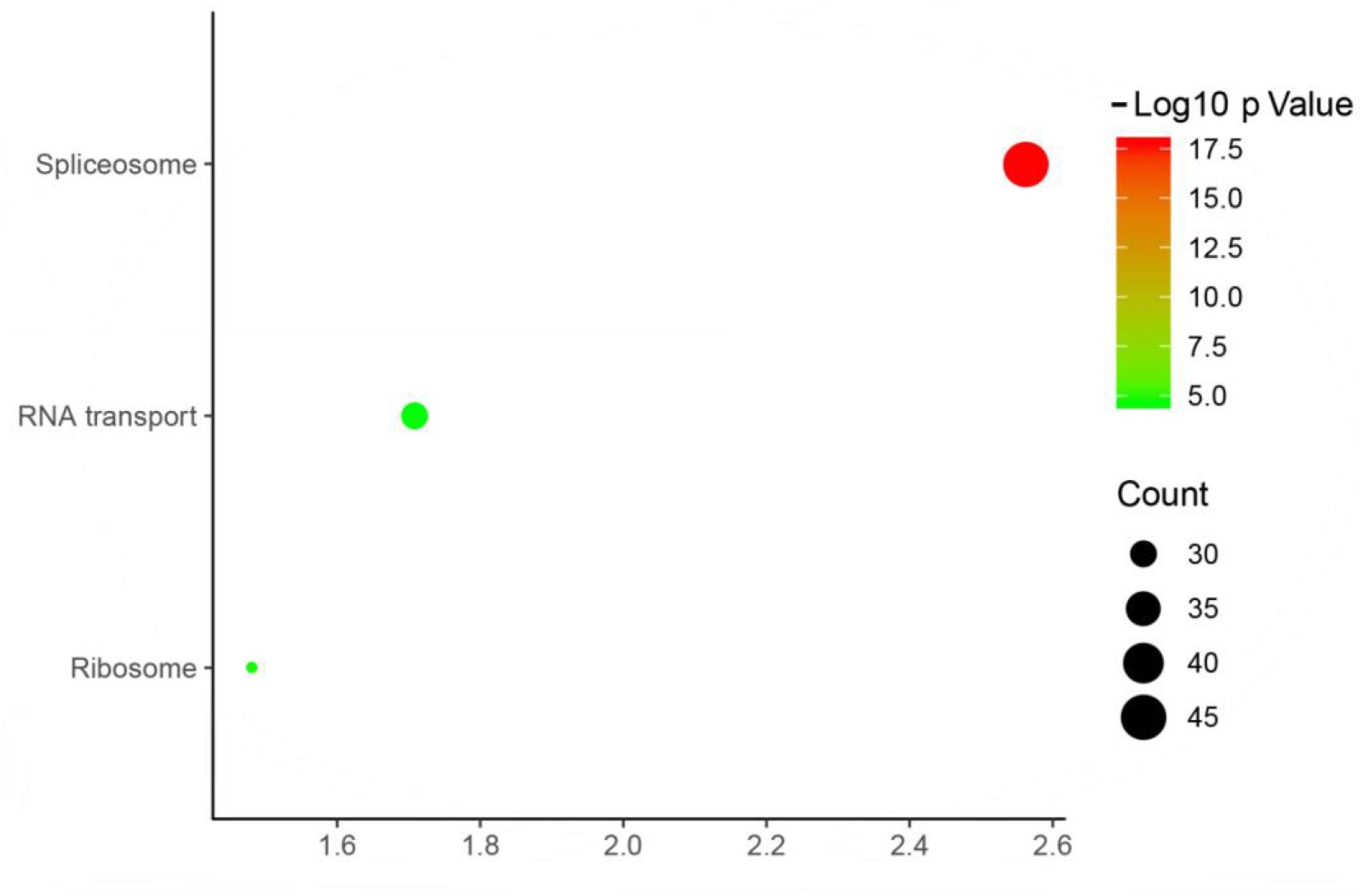 Fig5. KEEG Biological Pathway Enrichment Analysis.
Fig5. KEEG Biological Pathway Enrichment Analysis.
GO Enrichment Analysis:
- Differential genes were analyzed for GO enrichment in molecular function, biological process, and cellular component categories.
- Significant GO terms were identified, highlighting the functional roles of the binding sites.
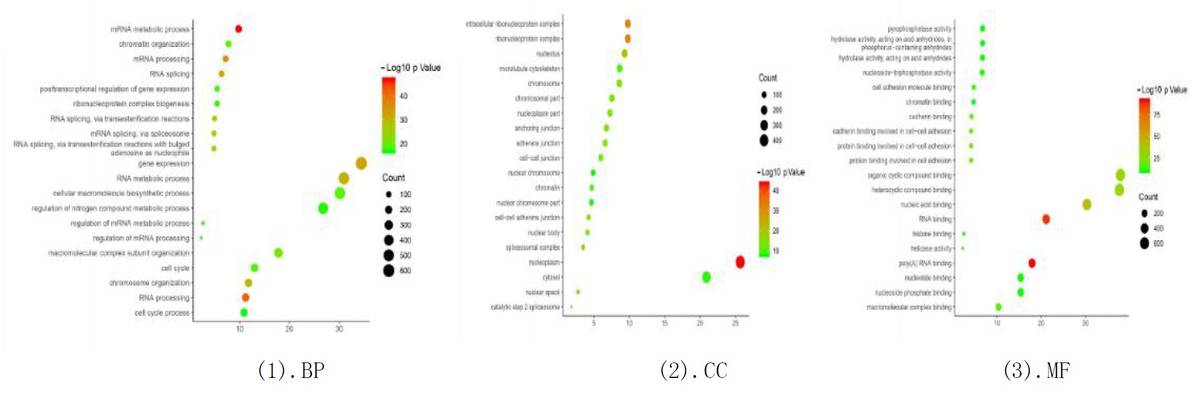 Fig6. GO Enrichment Analysis of Differential Genes.
Fig6. GO Enrichment Analysis of Differential Genes.
Motif Analysis:
- Consensus motifs were identified in the binding peaks, providing insights into the sequence preferences of X protein.
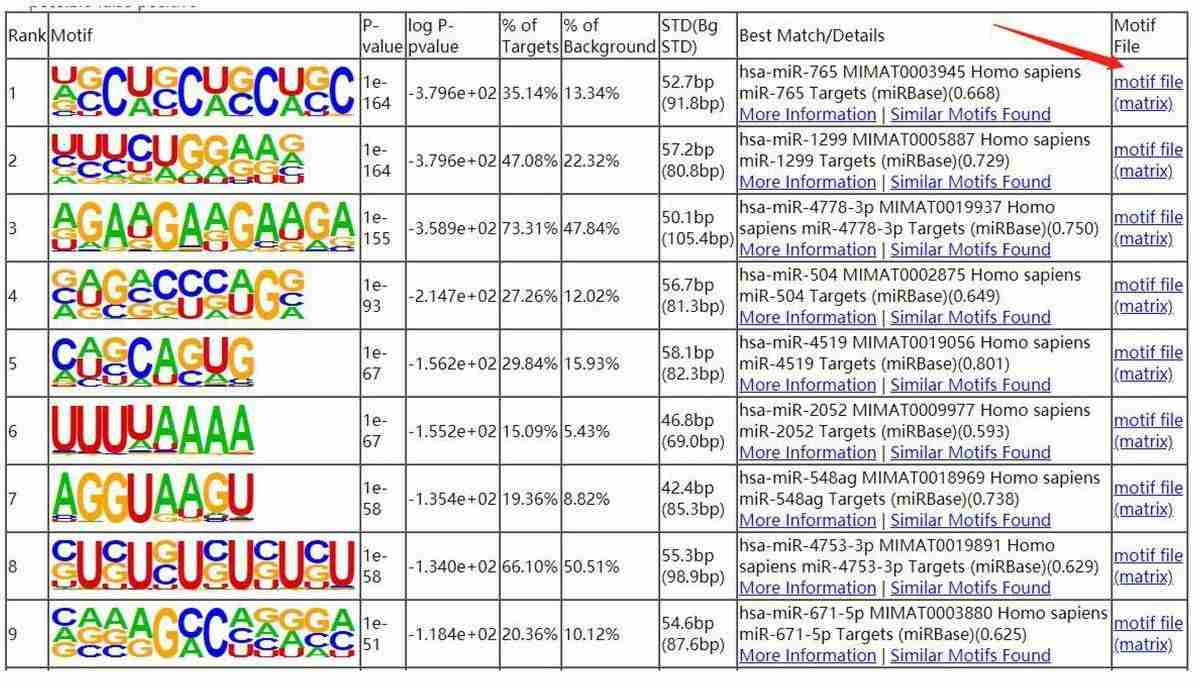 Fig7. The smaller the P value, the more significant of the motif.
Fig7. The smaller the P value, the more significant of the motif.
FAQs
Resources
References:
- Feng J.; et al. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7(9):1728-1740.
- Du C, Volkan P. Using Chromatin Immunoprecipitation (ChIP) to Study the Chromatin State in Drosophila. Cold Spring Harb Protoc. 2025;2025(1):pdb.top108139.