LBVS methods use compound information to predict activity, which measure the similarity of the compounds in the library to reference compounds that are active towards a target of interest. 2D or 3D chemical structures or molecular descriptors of the known actives are used to retrieve other (‘similar”) compounds of interest in a database using different types of similarity measures or by seeking a common substructure or pharmacophore between the query molecules and the scanned libraries. LBVS has the significant advantages over other virtual screening approaches because it does not involve macromolecules in the calculations and requires no prior knowledge of active ligands. It is chosen as one of the most popular approaches for drug discovery and lead optimization especially when three-dimensional structures of potential drug targets are not available.
Profacgen provides various LBVS methods including 2D molecular similarity approaches (fingerprint-based methods), 3D similarity searches (molecular shape colored or not by physicochemical properties, pharmacophore, and energy fields around the molecules such as electrostatic properties), machine learning and 2D/3D QSAR (quantitative structure-activity properties) modeling.
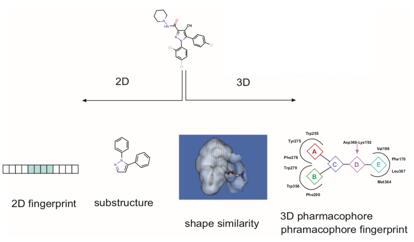
Figure 1. Ligand-based virtual screening methods. (Comb Chem High Throughput Screen. 20111)
LBVS similarity method is a very simple and computationally inexpensive method to retrieve compounds with similar characteristics to known ligands. In 2D molecular similarity methods, the molecular fingerprint of known ligands that bind to a target is used to find molecules with similar fingerprints in the electronic libraries, while in 3D similarity methods, the 3D shape of compounds in libraries is compared to the 3D shape of known active ligands, which are used as a reference.
Pharmacophore modeling has also been widely used in LBVS to generate predictive models suitable for the design of novel compounds. At Profacgen, we build a pharmacophore model by superposing a set of structurally diverse compounds that bind to the same target and extract common chemical features that are essential for their bioactivity. Potential ligand candidates can be then identified by using the pharmacophore model to virtually screen libraries of compounds.
Our advantages:
[1]Polgár, T.; Keseru, G. M., Integration of virtual and high throughput screening in lead discovery settings. Comb Chem High Throughput Screen 2011, 14 (10), 889-97.
Fill out this form and one of our experts will respond to you within one business day.