Recombinant protein expression in bacteria often results in the formation of both inactive and insoluble protein that accumulates as intracellularprotein aggregates called inclusion bodies [1]. It has beenshown that 70-80% of recombinant proteins expressed in E.coli are as inclusion bodies [2]. This is probably due to the independence of the protein type in bacterial systems. In cases of expression of eukaryotic proteins, which usually contain cysteines that are prone to form disulfide bonds in the nativestate, the bacterial system maynot support the appropriate pairing of disulfide bonds in the newly-produced protein thus leads to the presence of insoluble protein pellets [3].
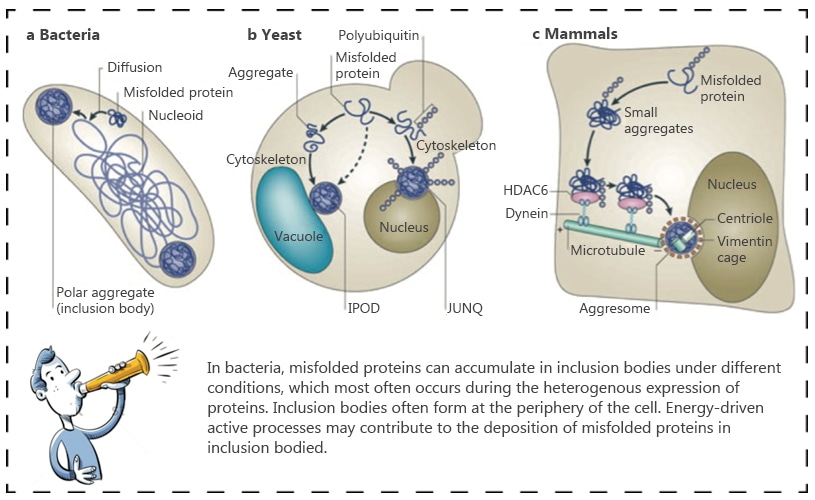
Inclusion bodies are not restricted to E.coli, they can also form in yeast, mammalian, and insect cells. Inclusion bodies recovered from cell lysates by low-speed centrifugation are heavily contaminated with E.coli cell wall and outer membranecomponents.
Here is what we do at Profacgen to obtain native and soluble protein form. First of all, the insoluble protein pellets must be separated from other cellular components by homogenization, washing and centrifugation; which is then followed by the refolding of protein by solubilization in denaturants, such as guanidine hydrochloride or urea [1]. Besides, certain reducing reagents are added to reduce the polypeptide cysteines to break existing disulfide bonds to obtain monomeric peptide chains [4].
Generally speaking, we apply selective extraction with detergents and low concentrations of urea or guanidine chloride to solublize the protein pellets. These basic steps can dissolve about 60% of the pellet protein. The challenge, therefore, is not to purify the recombinantly-derived protein, but to solubilize it and then fold it into native and biologically active protein [5].
Profacgen possesses expertise in recovery of correctly folded protein, which requires laborious and expensive processing of inclusion bodies by conventional methods. We guarantee the structural integrity and native conformation of your target protein. Here we provide the most common procedure used in our laboratory for your reference.
Inclusion bodies are noncrystalline, amorphous structures; however, there is evidence that the constituent densely packed proteins may have native-like secondary structures [6]. The decision of whether to work with insoluble recombinant protein or to put more effort into generating soluble protein (e.g., try to modify the expression vector, change the host strain and fermentation conditions or co-express with molecular chaperones etc.) depends on the characteristics of the protein.
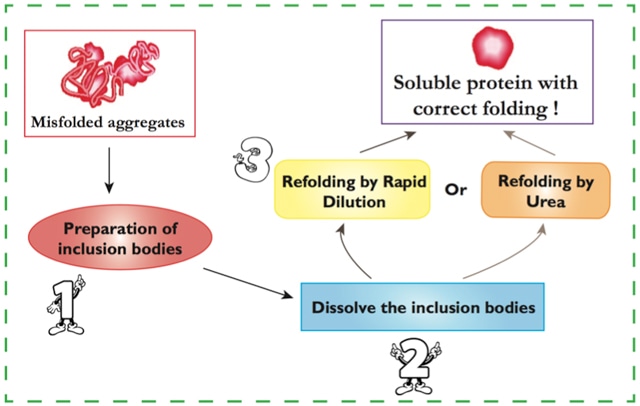
Step 1. Preparation of inclusion bodies:
| a. | Harvest bacteria after induction. |
| b. | Lyse bacteria by sonication in the buffer containing Tri-HCl, NaCl, EDTA, NaN3, Triton-X100, PMSF and DTT. 50 ml aliquot usually works well for sonication. |
| c. | Add MgSO4 to chelate the EDTA,then add DNase and lysozyme to the lysate and incubate at RT. |
| d. | Centrifuge the mixture and collect inclusion bodies. Crush the pellet and re-suspend by sonication in the lysis buffer. Repeat this step. |
| e. | Wash the inclusion bodies with lysis buffer without Triton-X100. Re-suspend the pellet by sonication. |
| f. | Eventually collect the inclusion body pellet by centrifugation. |
Step 2. Dissolve the inclusion bodies:
| a. | Add Tris buffer (with Glycine) into pure inclusion body. |
| b. | Disperse the pellets by sonication and dissolve the suspension dropwise. Stir vigorously in Tris buffer containing urea. |
| c. | Add GSSH (oxidized glutathione) and GSSG (reduced glutathione), and stir overnight. |
Step 3. Protein refolding:
| Method I: Refolding by urea. | |
| a. | Refolding buffers include Tris and L-Arginine supplemented with a concentration gradient Urea solution. |
| b. | Set the pH. |
| c. | Add EDTA, protease inhibitors and PMSF immediately before use. |
| d. | Dialyze against refolding buffer with concentration gradient urea solution. |
| e. | Dilute refolding buffer with water. |
| f. | Dialyze against running buffer with PMSF. |
| Method II: Refolding by rapid dilution. | |
| Add solubilized inclusion bodies dropwise into refolding buffer with rapid stirring. | |
Generally, a large portion of misfolded aggregates and multimers will crash out when the protein is refolded or concentrated. The yield by mass of refolded protein from a pellet for most proteins is about 2-5%, although some proteins refold more easily (about 20%).
Click here to contact us for more technical information.
[1]Fischer B, Sumner I, Goodenough P. Isolation, renaturation, and formation of disulfide bonds of eukaryotic proteins expressed in Escherichia coli as inclusion bodies[J]. Biotechnol Bioeng, 1993, 41(1): 3-13.
[2]Yang Z, Zhang L, Zhang Y, et al. Highly efficient production of soluble proteins from insoluble inclusion bodies by a two-step-denaturing and refolding method[J]. PLoS One, 2011, 6(7): e22981.
[3]Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation[J]. Nat Rev Mol Cell Biol, 2010, 11(11): 777-88.
[4]O'callaghan C A, Tormo J, Willcox B E, et al. Production, crystallization, and preliminary X-ray analysis of the human MHC class Ib molecule HLA-E[J]. Protein Sci, 1998, 7(5): 1264-6.
[5]Palmer I, Wingfield P T. Preparation and extraction of insoluble (inclusion-body) proteins from Escherichia coli[J]. Curr Protoc Protein Sci, 2004, Chapter 6: Unit 6 3.
[6]Oberg K, Chrunyk B A, Wetzel R, et al. Nativelike secondary structure in interleukin-1 beta inclusion bodies by attenuated total reflectance FTIR[J]. Biochemistry, 1994, 33(9): 2628-34.
Fill out this form and one of our experts will respond to you within one business day.