DAP-seq Service (DNA affinity purification sequencing)
Background
Introduction
ChIP-seq is a popular method that depends heavily on having the right antibodies for each target protein, which can be tough to get, particularly for proteins found in lesser-known organisms or in low amounts. The process itself is complex and time-consuming, often not producing the best quality data. DAP-seq (DNA Affinity Purification Sequencing) has emerged as a better alternative to tackle these issues effectively.
Advantages of DAP-seq (Compared to ChIP-seq, DAP-seq offers several key advantages)
- High Throughput: You can dig into binding sites for loads of transcription factors all at the same time. It's really efficient!
- No Antibody Requirement: No need to worry about finding specific antibodies, which makes it easier to work with different species and proteins.
- Retention of Modifications: It hangs on to those special tweaks in tissues or cell lines, so you get a real-world view of what's going on.
- Cost-Effective: Great for big projects that need to watch the budget – it won't cost you a fortune!
What is DAP-seq?
DAP-seq is a method that maps transcription factor binding sites across the genome without needing specific antibodies. It combines in vitro protein expression with high-throughput sequencing, making it more accessible and cheaper. By allowing expressed proteins to bind with DNA fragments, DAP-seq effectively identifies binding sites and is particularly useful for studying transcription factors in species like plants, where getting the right antibodies can be tough.
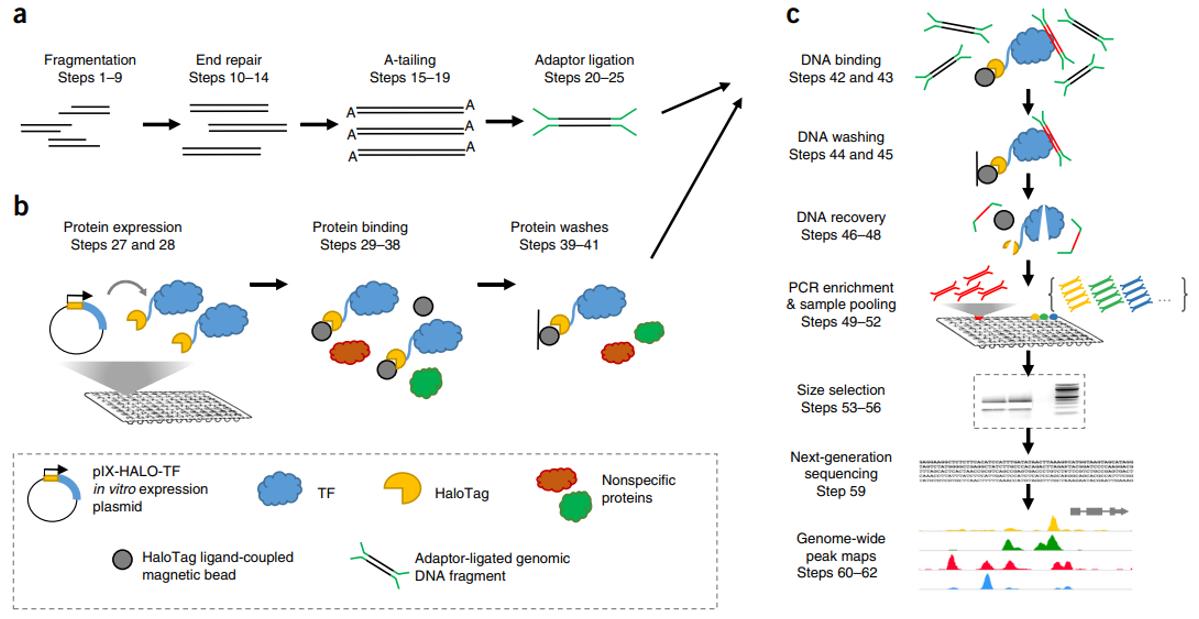 Fig1. DAP-seq protocol overview (Bartlett, A.; et al. 2017)
Fig1. DAP-seq protocol overview (Bartlett, A.; et al. 2017)
Applications of DAP-seq
- Finding Transcription Factor Spots: Use it to find where transcription factors hang out on the genome and figure out what they do.
- Checking Out Histone Changes: Look at different histone modifications at certain DNA spots and see how they mess with gene expression.
- Mapping Gene Networks: Make detailed maps of regulatory elements to get a grip on how gene networks are controlled.
- Understanding Disease: Dive into how transcription factors play a part in diseases by looking at weird binding sites and how they affect gene expression.
Service Procedure
As an expert in protein-nucleic acid interaction research, Profacgen's DAP-seq technology platform enables the high-throughput creation of genome-wide binding site maps and efficiently analyzes the effects of modifications in genomic DNA libraries on transcription factor binding.
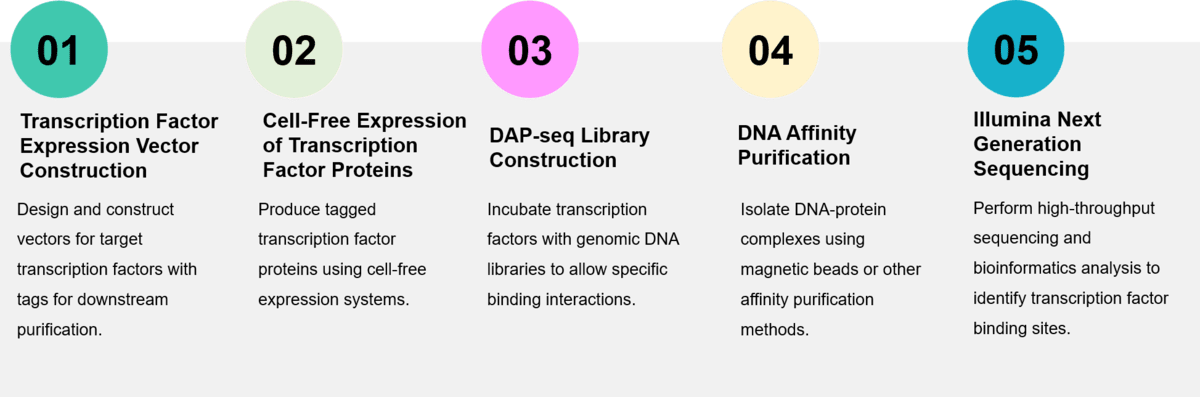
Deliverables
- High-throughput sequencing raw data
- Detailed bioinformatics analysis reports
- Transcription factor binding motif sequences
You only need to provide the transcription factor sequence and the genome sample of the material to be tested. Profacgen will complete the DAP-seq service for you, delivering comprehensive data and actionable insights.
Our DAP-seq service has got everything you need if you're looking into transcription factor binding sites, histone changes, or gene regulation networks. With our know-how and cutting-edge tech, you can count on us to give you top-notch results for your research.
Why Choose Profacgen?
- Expertise and Experience: The folks at Profacgen know their stuff when it comes to nucleic acid-protein interactions, so you can be sure of getting great results.
- High-Throughput and Efficient: Our setup lets you quickly and thoroughly map out transcription factor binding sites all over the genome.
- Customized Solutions: We can tweak our services to fit what you need, whether it's building vectors or doing bioinformatics analysis.
- Competitive Pricing: Our fair prices and deep know-how have made us a go-to partner for collaborators around the globe.
Case Study
Background
This project utilized DNA Affinity Purification Sequencing (DAP-seq) to study the binding sites of transcription factors in vitro. The goal is to identify the interaction DNA of target protein based on DAP-seq. Enriched DNA fragments were sequenced by Illumina sequencing (using NextSeq 500). Comprehensive data analysis was also included in our service.
Results
- Target CDS Sequence Amplification: The target transcription factor CDS sequence was successfully amplified, with the target size confirmed through agarose gel electrophoresis.
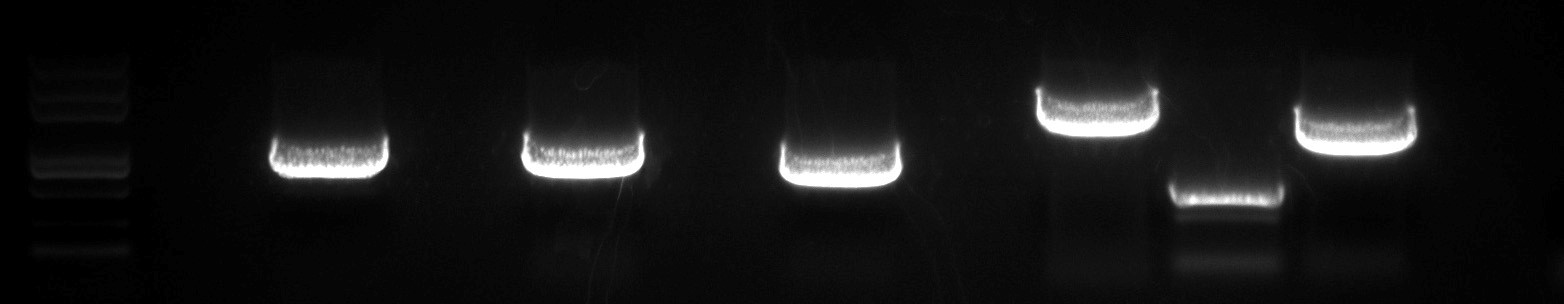 Fig2. Agarose Gel Electrophoresis of Amplified Target DNA Fragments.
Fig2. Agarose Gel Electrophoresis of Amplified Target DNA Fragments.
- Protein Expression Vector Construction: The correct protein expression vector was constructed and validated using colony PCR.
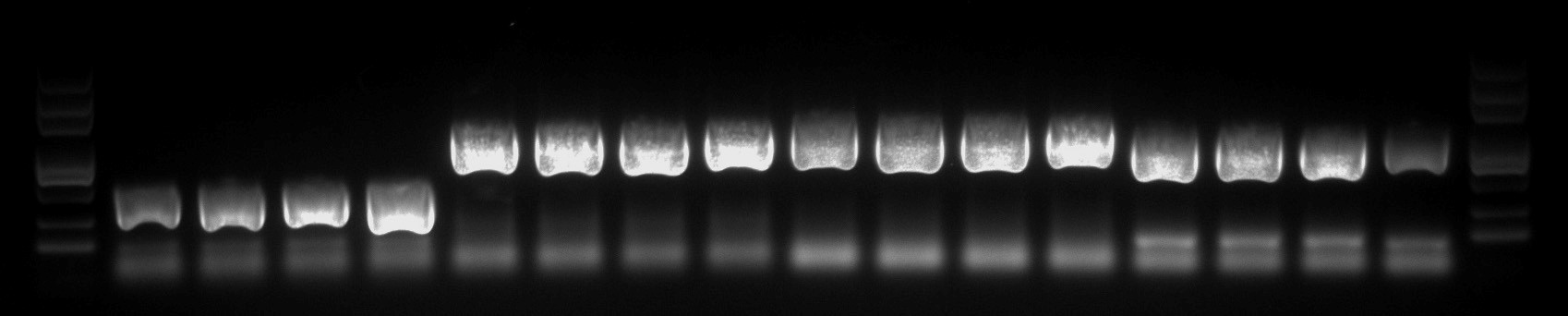 Fig3. Colony PCR Screening for Positive Protein Expression Vector.
Fig3. Colony PCR Screening for Positive Protein Expression Vector.
- In Vitro Protein Expression and Detection: The target transcription factor protein was successfully expressed in vitro and detected using Western Blot.
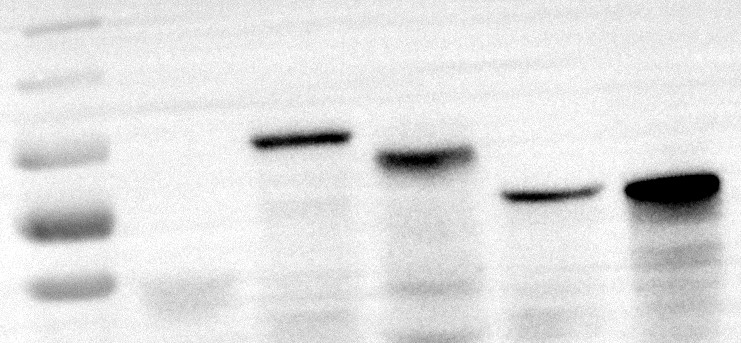 Fig4. Western Blot Analysis for Protein Expression.
Fig4. Western Blot Analysis for Protein Expression.
- DNA Affinity Purification Library: The DNA affinity purification library was constructed and validated.
 Fig5. Agarose Gel Electrophoresis of DNA Affinity Purification Library.
Fig5. Agarose Gel Electrophoresis of DNA Affinity Purification Library.
- Affinity Purification of Proteins and Libraries: Specific binding of transcription factors to genomic DNA was confirmed.
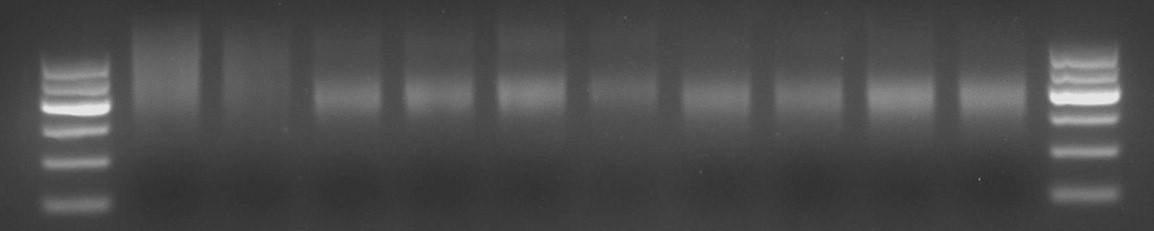 Fig6. Agarose Gel Electrophoresis of after Protein and DNA Affinity.
Fig6. Agarose Gel Electrophoresis of after Protein and DNA Affinity.
- Data Analysis: Comprehensive analysis provided detailed insights into transcription factor binding sites, peak distribution, and functional annotations. Key findings included:
- Peak Distribution in Genome Functional Regions: Genome-wide functional regions are divided into Promoter, Exon, Intron, TTS and Intergenic regions. After the peak is annotated, the results of the annotation and the distribution of the peak are counted.
- Gene Ontology Analysis: Enriched functions related to transcription factor binding were identified.
- KEGG Analysis: Significant pathways associated with binding sites were identified.
- Motif Analysis: Conserved motifs in peak regions were identified, providing insights into transcription factor binding preferences.
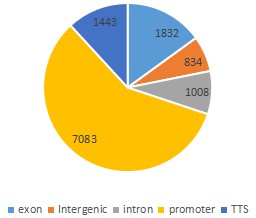 Fig7. Peak Distribution in Genome Functional Regions.
Fig7. Peak Distribution in Genome Functional Regions.
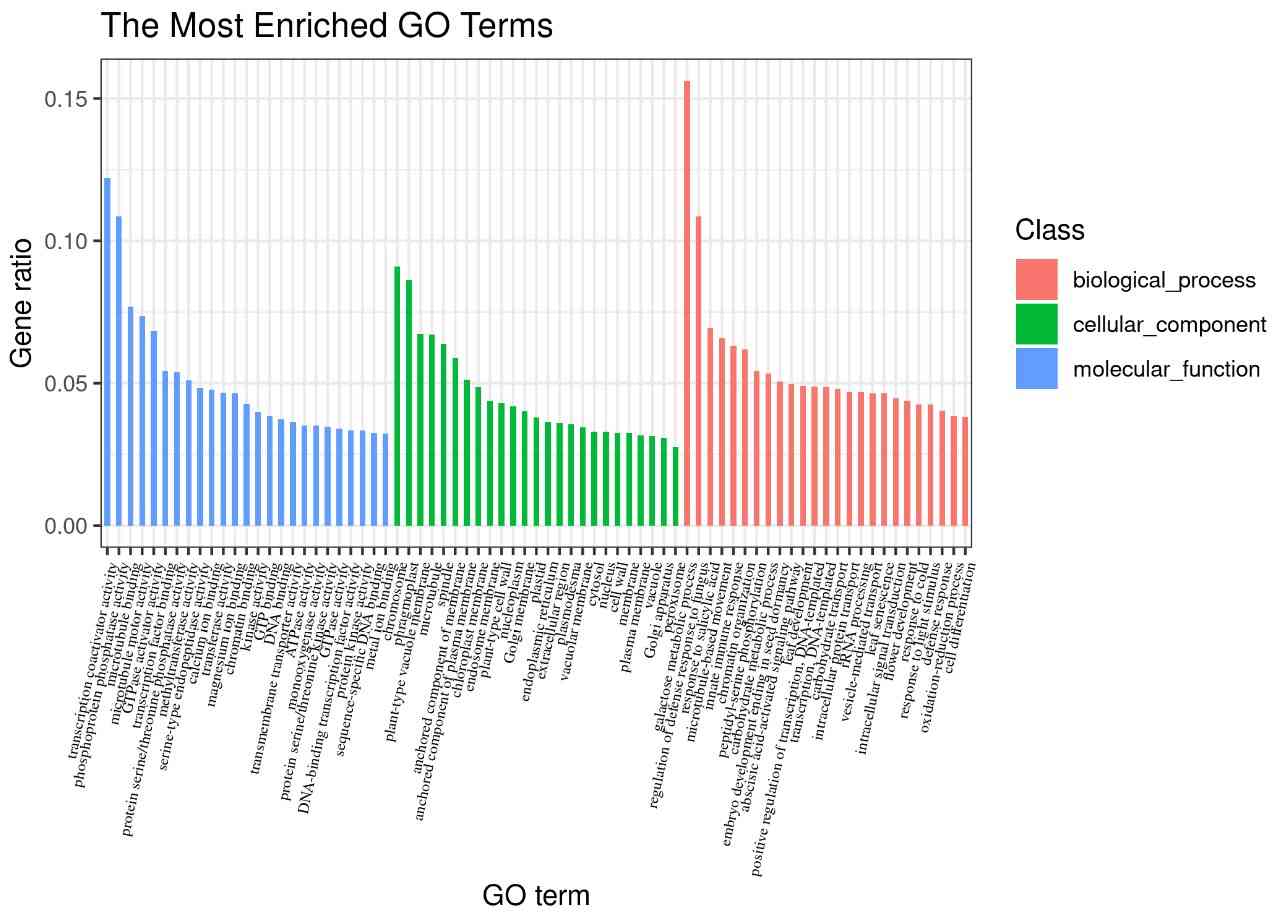 Fig8. GO Enrichment Bar Graph of Peak Associated Genes.
Fig8. GO Enrichment Bar Graph of Peak Associated Genes.
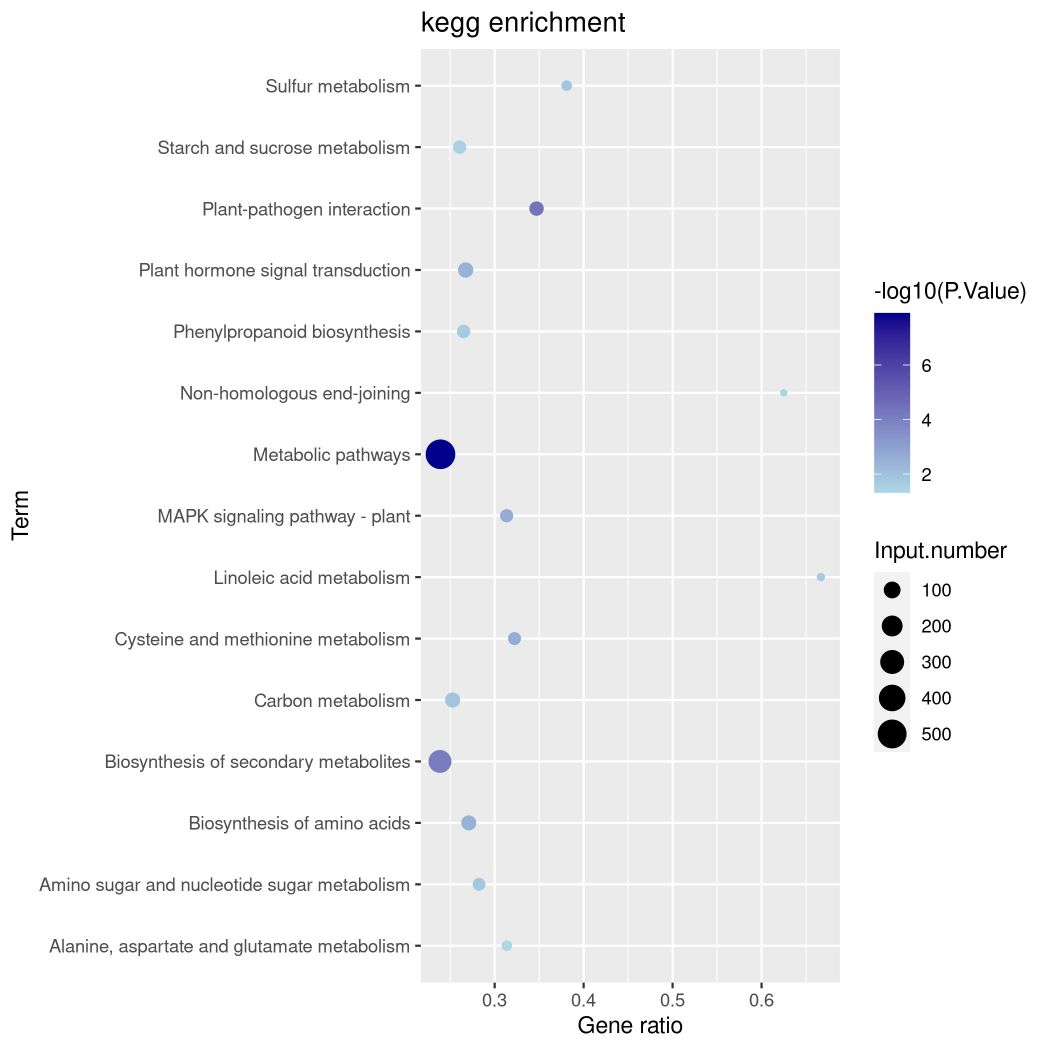 Fig9. Bubble Diagram of Enrichment of Promoter Related Genes Top15 KEGG.
Fig9. Bubble Diagram of Enrichment of Promoter Related Genes Top15 KEGG.
FAQs
Resources
| Chromatin Immunoprecipitation (ChIP) Assay Service | ChIRP-Seq Service | miCLIP-seq | DNase I Footprinting Assay Service | Electrophoretic Mobility Shift Assay (EMSA) Service |
References:
- Bartlett A.; et al. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc. 2017;12(8):1659-1672.
- Hutin S.; et al. Identification of Plant Transcription Factor DNA-Binding Sites Using seq-DAP-seq. Methods Mol Biol. 2023;2698:119-145.