Antibody Modeling
Antibodies are proteins that play a critical role in our immune system for recognizing various antigens. Antibodies are increasingly used as diagnostic tools and therapeutic drugs, and an important component of the antibody engineering process is the availability of high quality three-dimensional structures. Experimental determination of antibody structures is laborious and not always possible. On the contrary, computational modeling provides a fast and inexpensive route for generating antibody structural models.
The variable domains (Fvs) of the heavy and light chain are of special interest in structural modeling, as they typically impart most or all of the specificity of an antibody for its antigen target. Modeling of the Fv region of an antibody involves prediction of both the framework (FR) region and the variable loop (CDR) region. Our protocol starts with a template search for the framework region (FR) in our curated antibody database, which is derived from the publically available crystal structures in the Protein Data Bank (PDB). After that, templates for the CDR region are chosen based on homology from canonical loop conformations that are clustered in the database. Structural models for the Fv region are then built along with refinement of the H3 loop. It is also possible to model the entire antibody including the constant region (Fc).
Workflow for predicting the structure of antibodies
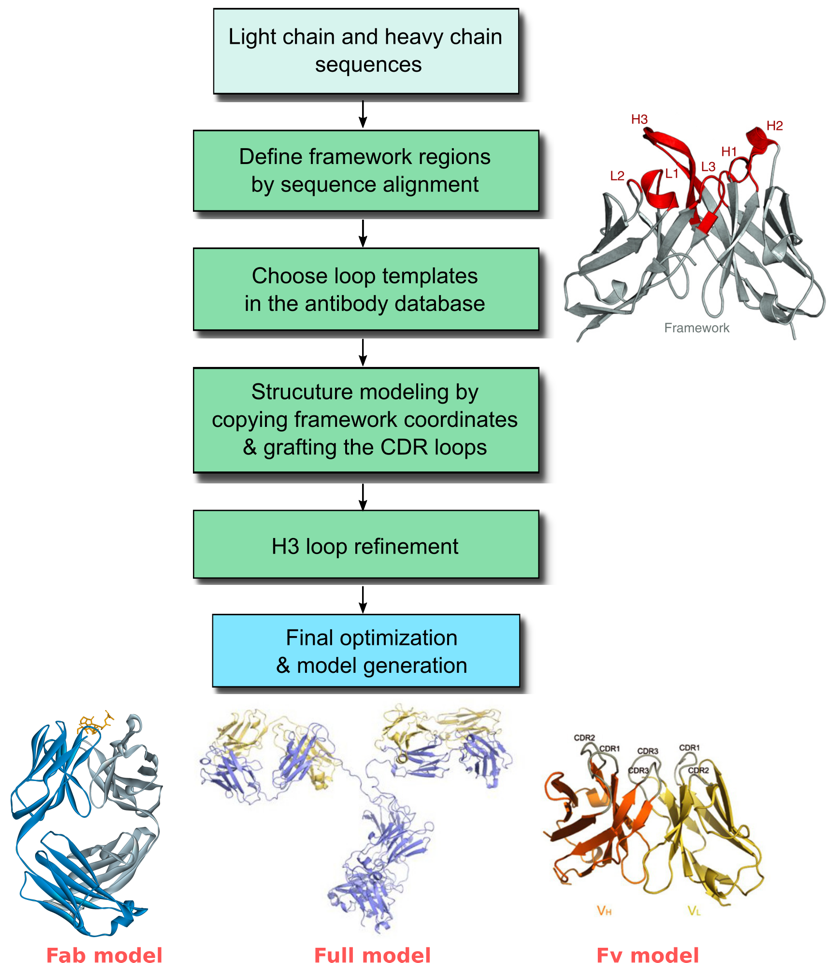
Profacgen takes advantage of computational modeling method to help customers predict the three-dimensional structure of antibodies of interest. We have extensive experiences with the structural modeling of various antibodies. The resultant antibody models are all quality verified and can be used for designing and engineering novel antibodies with desired therapeutic properties.
Features
High quality 3D Fv, Fab or full antibody modelCurated antibody databases obtained from the PDB
Selection of separate templates for the heavy and light chains
CDR H3 loop modeling and refinement
Inclusion of antigen, ligands, cofactors, and water in the model
Compatible with common antibody numbering schemes: Chothia, Kabat, IMGT, AHo
Prediction of physical properties
Model-based prediction for antibody humanization
We customize the service according to the specific requirements from the customers. Please feel free to contact us for more details about our antibody modeling service.